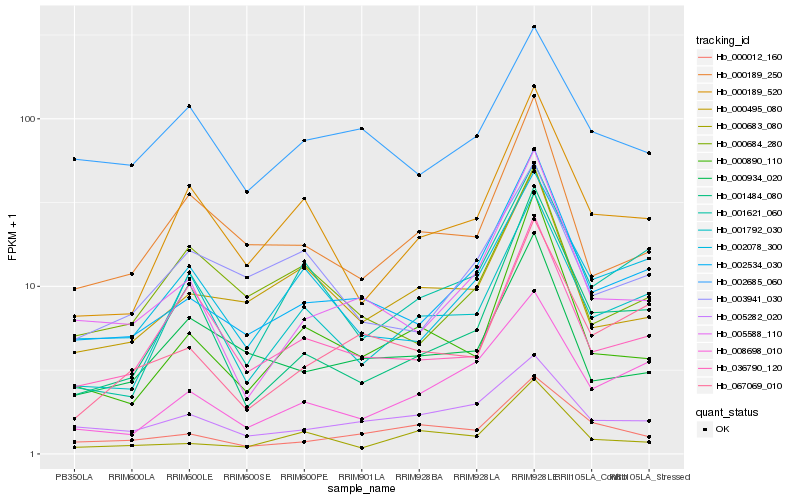
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002534_030 |
0.0 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 2 |
Hb_005588_110 |
0.1010856329 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 3 |
Hb_008698_010 |
0.1250518105 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_067069_010 |
0.1260491447 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 5 |
Hb_002078_300 |
0.1341570172 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001484_080 |
0.1524848237 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 7 |
Hb_000890_110 |
0.1527844502 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 8 |
Hb_000189_520 |
0.1538013435 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 9 |
Hb_000495_080 |
0.1538969505 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 10 |
Hb_000189_250 |
0.1551379954 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_005282_020 |
0.1551975198 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 12 |
Hb_001621_060 |
0.1577932151 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000934_020 |
0.1581405328 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |
| 14 |
Hb_001792_030 |
0.1586140146 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 15 |
Hb_000012_160 |
0.1609234178 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 16 |
Hb_036790_120 |
0.1613517712 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003941_030 |
0.1633102851 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 18 |
Hb_000683_080 |
0.1645966641 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 19 |
Hb_002685_060 |
0.1660269286 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 20 |
Hb_000684_280 |
0.1667733229 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g09650, chloroplastic [Jatropha curcas] |