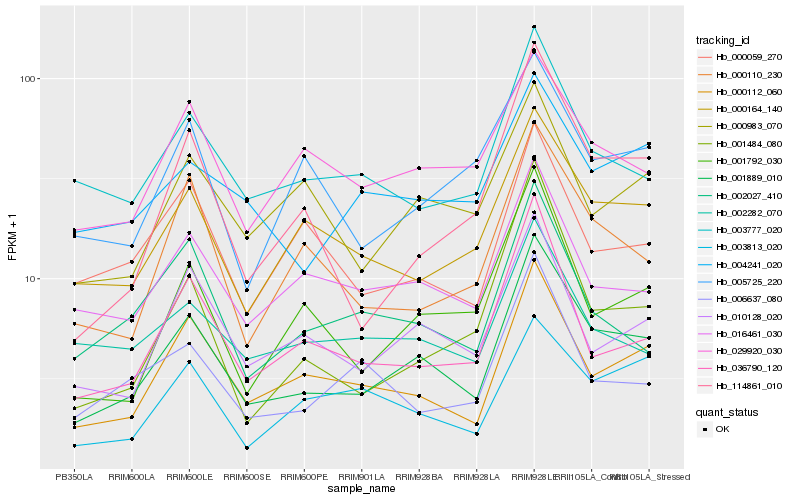
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001889_010 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: protein CHLOROPLAST IMPORT APPARATUS 2 [Jatropha curcas] |
| 2 |
Hb_000112_060 |
0.1156919832 |
- |
- |
PREDICTED: thioredoxin-like protein CITRX, chloroplastic [Jatropha curcas] |
| 3 |
Hb_010128_020 |
0.1271221645 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 4 |
Hb_000164_140 |
0.1285625891 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 5 |
Hb_036790_120 |
0.1329565136 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001792_030 |
0.1345277993 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 7 |
Hb_114861_010 |
0.1373429442 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 8 |
Hb_000110_230 |
0.1408273496 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_016461_030 |
0.1409924485 |
- |
- |
PREDICTED: putative leucine--tRNA ligase, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_003777_020 |
0.1412825927 |
- |
- |
Fructose-1,6-bisphosphatase, cytosolic, putative [Ricinus communis] |
| 11 |
Hb_000983_070 |
0.1447480866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_029920_030 |
0.1456042363 |
- |
- |
PREDICTED: 2-Cys peroxiredoxin BAS1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004241_020 |
0.148873865 |
- |
- |
PREDICTED: DAG protein, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_001484_080 |
0.1520381281 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 15 |
Hb_002027_410 |
0.1528214326 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000059_270 |
0.1534500473 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 17 |
Hb_005725_220 |
0.154558925 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 18 |
Hb_003813_020 |
0.1555913019 |
- |
- |
PREDICTED: uncharacterized protein LOC105638736 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006637_080 |
0.156906929 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002282_070 |
0.157126649 |
- |
- |
PREDICTED: protein DJ-1 homolog C isoform X2 [Jatropha curcas] |