| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
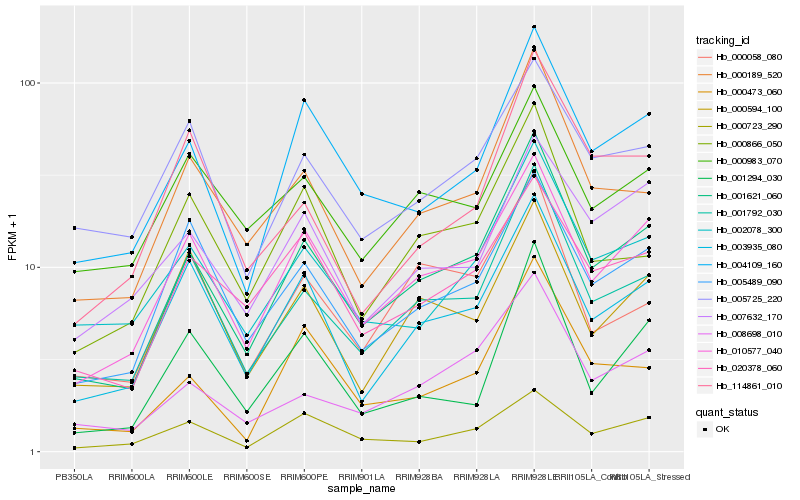
Hb_001621_060 |
0.0 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001792_030 |
0.0884310923 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 3 |
Hb_010577_040 |
0.0921369253 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 4 |
Hb_000189_520 |
0.1055989455 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 5 |
Hb_004109_160 |
0.1059572087 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_008698_010 |
0.1071064705 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 7 |
Hb_003935_080 |
0.1119167943 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002078_300 |
0.1161605569 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000473_060 |
0.1185582661 |
- |
- |
PREDICTED: psbP domain-containing protein 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005489_090 |
0.1307321388 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_020378_060 |
0.130922613 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 12 |
Hb_000866_050 |
0.1362456186 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 13 |
Hb_000723_290 |
0.1416340839 |
- |
- |
calcium lipid binding protein, putative [Ricinus communis] |
| 14 |
Hb_007632_170 |
0.1421277643 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 15 |
Hb_001294_030 |
0.1437541705 |
- |
- |
PREDICTED: uncharacterized protein LOC105644307 [Jatropha curcas] |
| 16 |
Hb_000058_080 |
0.1446787913 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 17 |
Hb_005725_220 |
0.1465485166 |
- |
- |
PREDICTED: uncharacterized protein LOC105630808 [Jatropha curcas] |
| 18 |
Hb_000594_100 |
0.147719084 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 19 |
Hb_114861_010 |
0.1509795695 |
- |
- |
STRESS ENHANCED protein 1 [Populus trichocarpa] |
| 20 |
Hb_000983_070 |
0.1533564609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |