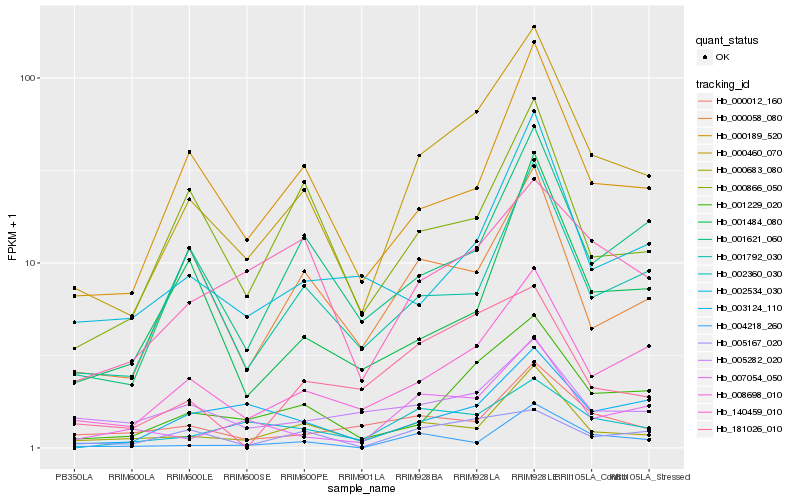
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000460_070 |
0.0 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 2 |
Hb_008698_010 |
0.1240429022 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 3 |
Hb_001229_020 |
0.1278716671 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_000683_080 |
0.1614842303 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 5 |
Hb_001621_060 |
0.1694804074 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000189_520 |
0.172112491 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 7 |
Hb_007054_050 |
0.1839670284 |
- |
- |
- |
| 8 |
Hb_001792_030 |
0.1867881126 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 9 |
Hb_002534_030 |
0.1876688995 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_000058_080 |
0.18874161 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 11 |
Hb_003124_110 |
0.1980546306 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 12 |
Hb_004218_260 |
0.1985919162 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 13 |
Hb_000866_050 |
0.200827588 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 14 |
Hb_000012_160 |
0.2017004307 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005167_020 |
0.203000046 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 16 |
Hb_002360_030 |
0.2054746046 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 17 |
Hb_181026_010 |
0.2058068984 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 18 |
Hb_001484_080 |
0.2069771213 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 19 |
Hb_140459_010 |
0.2098719365 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 20 |
Hb_005282_020 |
0.2105784185 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |