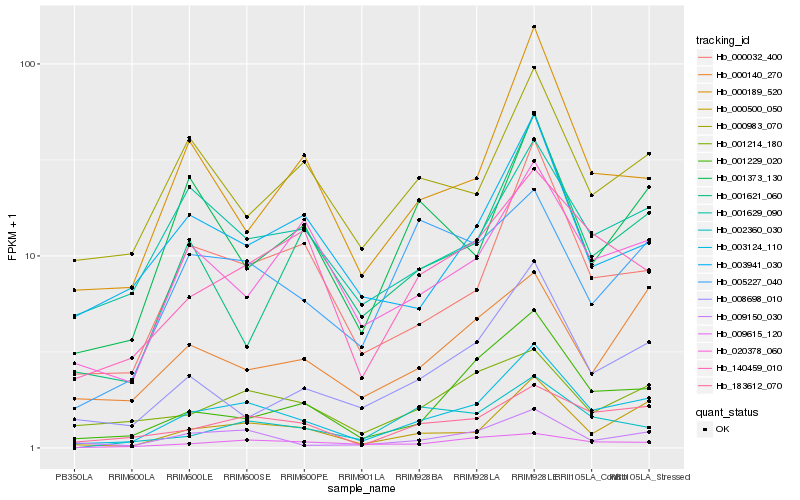
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003124_110 |
0.0 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 2 |
Hb_000500_050 |
0.136266177 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 3 |
Hb_183612_070 |
0.1399184283 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000032_400 |
0.1487204316 |
- |
- |
PREDICTED: UDP-glucose 4-epimerase GEPI48 [Jatropha curcas] |
| 5 |
Hb_009150_030 |
0.150241309 |
- |
- |
hypothetical protein POPTR_0010s14920g [Populus trichocarpa] |
| 6 |
Hb_001229_020 |
0.1612691106 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_140459_010 |
0.1642837107 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 8 |
Hb_008698_010 |
0.1743772438 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 9 |
Hb_020378_060 |
0.1763361127 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 10 |
Hb_000189_520 |
0.176868791 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 11 |
Hb_001629_090 |
0.1775596213 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001214_180 |
0.1788457555 |
- |
- |
- |
| 13 |
Hb_003941_030 |
0.1836904148 |
- |
- |
Protease ecfE, putative [Ricinus communis] |
| 14 |
Hb_009615_120 |
0.1846859542 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 15 |
Hb_001621_060 |
0.1876669687 |
- |
- |
PREDICTED: thylakoid lumenal 15 kDa protein 1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000983_070 |
0.1884000463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005227_040 |
0.1897515906 |
transcription factor |
TF Family: HB |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000140_270 |
0.190347494 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 19 |
Hb_002360_030 |
0.191036556 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 20 |
Hb_001373_130 |
0.1910713229 |
- |
- |
conserved hypothetical protein [Ricinus communis] |