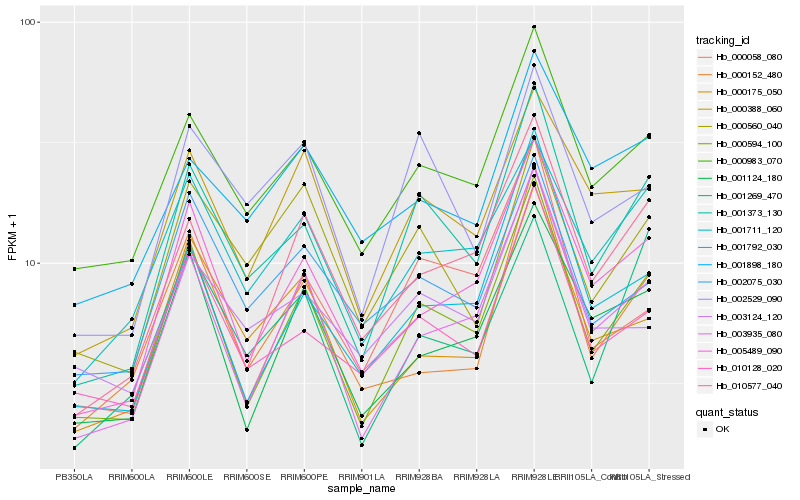
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001373_130 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000594_100 |
0.0792685161 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000983_070 |
0.101535984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010128_020 |
0.1133785824 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 5 |
Hb_002529_090 |
0.1184104457 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_005489_090 |
0.1192284603 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 7 |
Hb_003935_080 |
0.1226003546 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001711_120 |
0.1287616844 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_010577_040 |
0.1288865001 |
- |
- |
PREDICTED: uncharacterized protein LOC105643738 [Jatropha curcas] |
| 10 |
Hb_001792_030 |
0.1297144474 |
- |
- |
PREDICTED: aminoacyl tRNA synthase complex-interacting multifunctional protein 1 [Jatropha curcas] |
| 11 |
Hb_000388_060 |
0.1338154717 |
- |
- |
fructokinase [Manihot esculenta] |
| 12 |
Hb_002075_030 |
0.1420432982 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000058_080 |
0.1427659833 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 14 |
Hb_000560_040 |
0.1432930837 |
- |
- |
PREDICTED: membrane-associated 30 kDa protein, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001269_470 |
0.1440714109 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 16 |
Hb_000175_050 |
0.1452823478 |
- |
- |
- |
| 17 |
Hb_003124_120 |
0.1466018114 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 18 |
Hb_001898_180 |
0.1479679656 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000152_480 |
0.1489945323 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001124_180 |
0.1498476311 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |