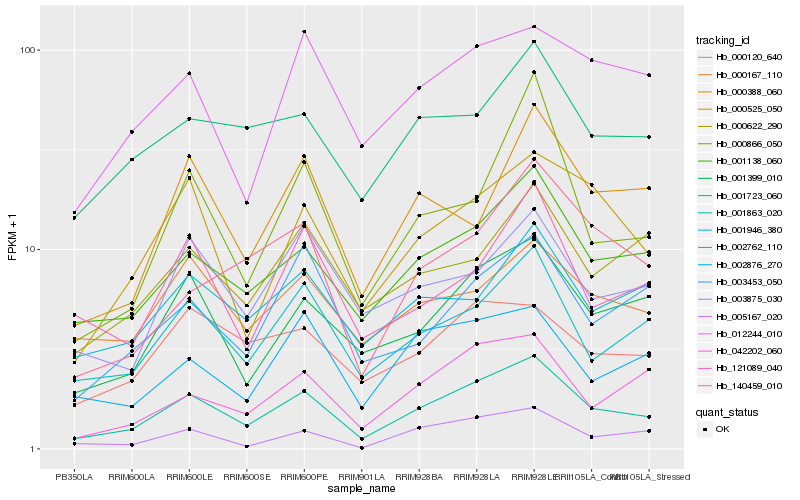
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_020 |
0.0 |
- |
- |
PREDICTED: protein FAM91A1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001138_060 |
0.1096333284 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 3 |
Hb_001946_380 |
0.1173779627 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Jatropha curcas] |
| 4 |
Hb_001399_010 |
0.12072848 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000525_050 |
0.1323852999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_042202_060 |
0.1363893814 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Jatropha curcas] |
| 7 |
Hb_005167_020 |
0.1401736899 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 8 |
Hb_001723_060 |
0.144933881 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 9 |
Hb_003453_050 |
0.1464075801 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 10 |
Hb_002876_270 |
0.1478691348 |
- |
- |
PREDICTED: probable magnesium transporter NIPA4 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002762_110 |
0.1501939871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_121089_040 |
0.1504291164 |
- |
- |
PREDICTED: GDSL esterase/lipase At4g10955 [Jatropha curcas] |
| 13 |
Hb_012244_010 |
0.1507782404 |
- |
- |
PREDICTED: uncharacterized protein LOC105641034 [Jatropha curcas] |
| 14 |
Hb_000622_290 |
0.1514297113 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 15 |
Hb_003875_030 |
0.1518067744 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 16 |
Hb_000388_060 |
0.1526227068 |
- |
- |
fructokinase [Manihot esculenta] |
| 17 |
Hb_000167_110 |
0.1534714522 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000120_640 |
0.1538395249 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000866_050 |
0.1538982403 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 20 |
Hb_140459_010 |
0.1539389781 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |