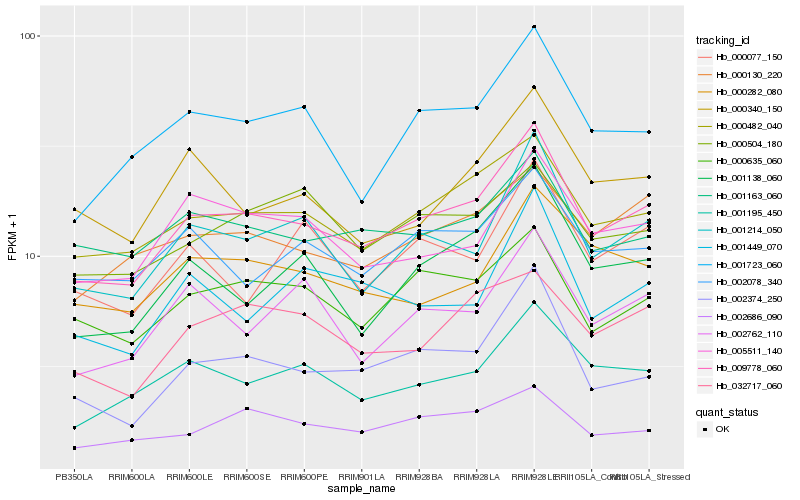
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009778_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 2 |
Hb_000482_040 |
0.0675581058 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 3 |
Hb_001214_050 |
0.0786351591 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001195_450 |
0.0837537141 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 5 |
Hb_001723_060 |
0.0848855147 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 6 |
Hb_002374_250 |
0.0851663499 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000635_060 |
0.0898432145 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 8 |
Hb_000130_220 |
0.0919215722 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_001138_060 |
0.09243713 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 10 |
Hb_002078_340 |
0.0943897014 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001163_060 |
0.0946414778 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_005511_140 |
0.0947925827 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 13 |
Hb_002686_090 |
0.0966546678 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 14 |
Hb_032717_060 |
0.0987113898 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ18 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000282_080 |
0.1005332682 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002762_110 |
0.1005554372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000340_150 |
0.1034426933 |
- |
- |
hypothetical protein JCGZ_21975 [Jatropha curcas] |
| 18 |
Hb_000504_180 |
0.1072278527 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 19 |
Hb_001449_070 |
0.1072847695 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000077_150 |
0.1077705271 |
- |
- |
PREDICTED: riboflavin synthase [Jatropha curcas] |