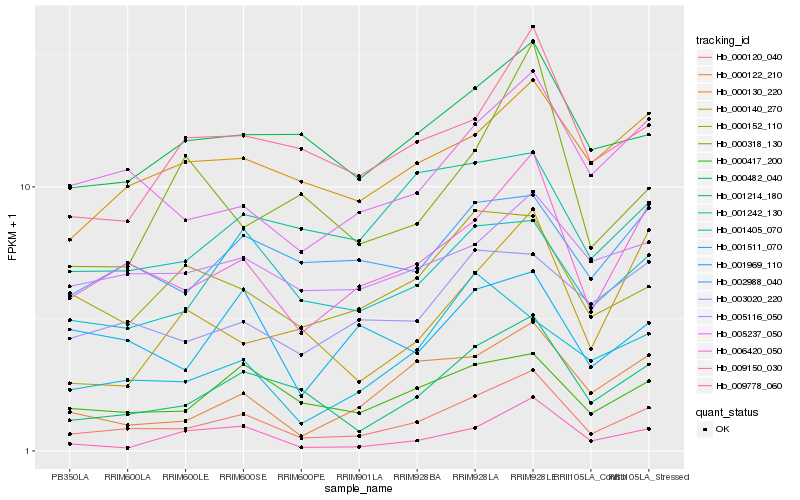
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_006420_050 |
0.0898361138 |
- |
- |
helicase, putative [Ricinus communis] |
| 3 |
Hb_001214_180 |
0.108564693 |
- |
- |
- |
| 4 |
Hb_000417_200 |
0.1380468842 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005237_050 |
0.1392762027 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630924 [Jatropha curcas] |
| 6 |
Hb_000122_210 |
0.1466901904 |
- |
- |
- |
| 7 |
Hb_009778_060 |
0.1496726811 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 8 |
Hb_001405_070 |
0.1504493286 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 9 |
Hb_000482_040 |
0.1550205415 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 10 |
Hb_000130_220 |
0.1594011726 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_000152_110 |
0.1607305157 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X1 [Jatropha curcas] |
| 12 |
Hb_009150_030 |
0.161891607 |
- |
- |
hypothetical protein POPTR_0010s14920g [Populus trichocarpa] |
| 13 |
Hb_003020_220 |
0.1635106413 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000140_270 |
0.1672886934 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 15 |
Hb_001969_110 |
0.1689676197 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002988_040 |
0.1690357894 |
- |
- |
- |
| 17 |
Hb_000318_130 |
0.1690992435 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_005116_050 |
0.1692287685 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_001511_070 |
0.1697015949 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 20 |
Hb_001242_130 |
0.1699470157 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |