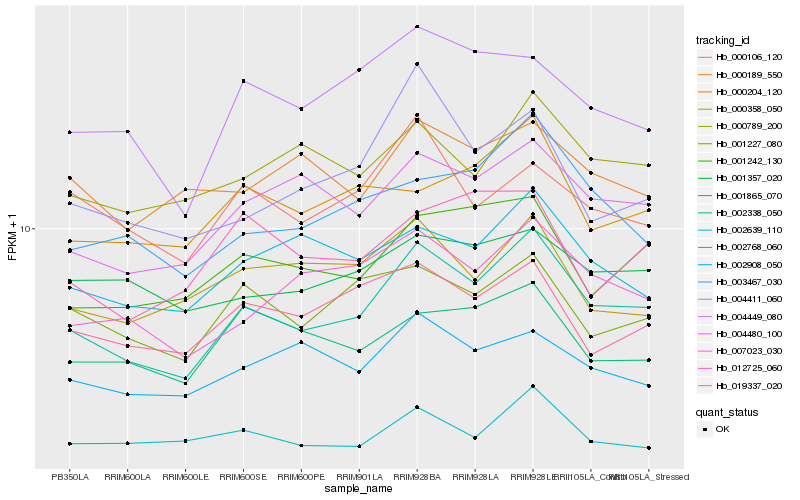
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002338_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105118489 isoform X1 [Populus euphratica] |
| 2 |
Hb_004480_100 |
0.0954067358 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 3 |
Hb_007023_030 |
0.098533396 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20710, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_003467_030 |
0.1052579501 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 5 |
Hb_001242_130 |
0.1064062804 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 6 |
Hb_002768_060 |
0.1106302998 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000789_200 |
0.1111956372 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 8 |
Hb_001227_080 |
0.1129359352 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_019337_020 |
0.1140251003 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 10 |
Hb_004449_080 |
0.1152801072 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 11 |
Hb_002908_050 |
0.1175605585 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 12 |
Hb_002639_110 |
0.1177432357 |
- |
- |
hypothetical protein L484_011673 [Morus notabilis] |
| 13 |
Hb_000204_120 |
0.1202044439 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 14 |
Hb_004411_060 |
0.1202297192 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 15 |
Hb_001865_070 |
0.1211506599 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001357_020 |
0.1220220462 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000358_050 |
0.1232641073 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 18 |
Hb_000106_120 |
0.1236881736 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 19 |
Hb_000189_550 |
0.1261630753 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 20 |
Hb_012725_060 |
0.1269743634 |
- |
- |
protein binding protein, putative [Ricinus communis] |