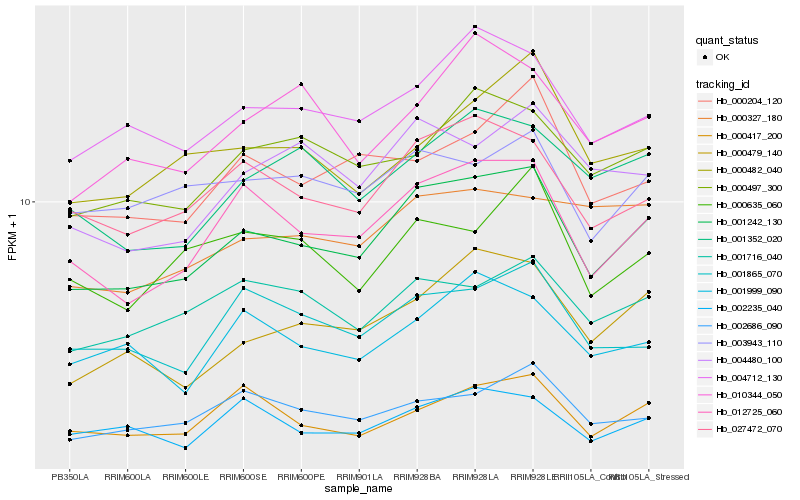
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001242_130 |
0.0 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 2 |
Hb_012725_060 |
0.0599542536 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_027472_070 |
0.0717591089 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 4 |
Hb_004712_130 |
0.0782462264 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 5 |
Hb_000479_140 |
0.085727598 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 6 |
Hb_000497_300 |
0.0886454336 |
- |
- |
peroxisomal membrane protein [Hevea brasiliensis] |
| 7 |
Hb_001352_020 |
0.0895105465 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_002686_090 |
0.0898110453 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 9 |
Hb_001716_040 |
0.0912885849 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 10 |
Hb_000204_120 |
0.0929363107 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 11 |
Hb_001865_070 |
0.092993631 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 12 |
Hb_004480_100 |
0.0938133629 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 13 |
Hb_003943_110 |
0.096018659 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 14 |
Hb_000417_200 |
0.0970657614 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002235_040 |
0.0970972763 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 16 |
Hb_010344_050 |
0.0971284998 |
- |
- |
Adagio 1 -like protein [Gossypium arboreum] |
| 17 |
Hb_000482_040 |
0.0971856775 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 18 |
Hb_001999_090 |
0.0976141728 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000635_060 |
0.0976325342 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 20 |
Hb_000327_180 |
0.100813609 |
- |
- |
PREDICTED: TBC1 domain family member 10B-like isoform X1 [Jatropha curcas] |