| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
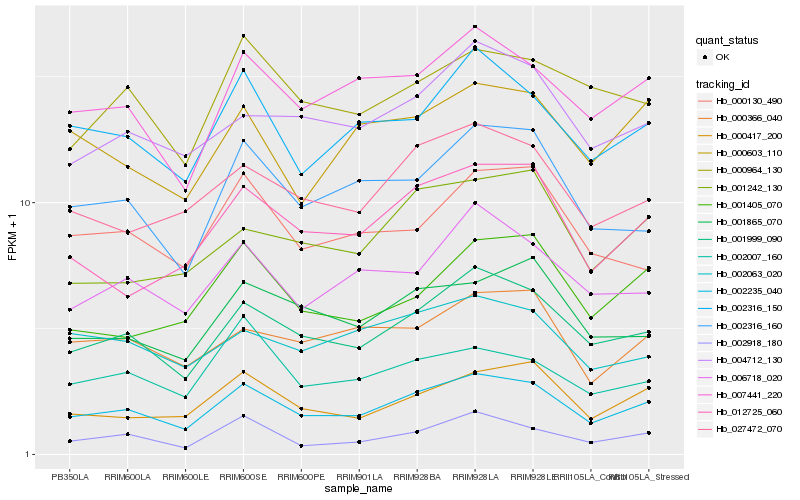
Hb_002235_040 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 2 |
Hb_001999_090 |
0.0506791644 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007441_220 |
0.0772746746 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 4 |
Hb_002316_150 |
0.0792174898 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640420 [Jatropha curcas] |
| 5 |
Hb_000417_200 |
0.0797596434 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002316_160 |
0.084733153 |
transcription factor |
TF Family: IWS1 |
PREDICTED: uncharacterized protein LOC105640422 [Jatropha curcas] |
| 7 |
Hb_012725_060 |
0.0854456602 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_027472_070 |
0.0857041371 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 9 |
Hb_006718_020 |
0.0887945749 |
- |
- |
PREDICTED: uncharacterized protein LOC105649330 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001865_070 |
0.0923320929 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004712_130 |
0.0924563841 |
- |
- |
PREDICTED: uncharacterized protein LOC105633074 [Jatropha curcas] |
| 12 |
Hb_001405_070 |
0.0930832794 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 13 |
Hb_000366_040 |
0.0957069601 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 14 |
Hb_001242_130 |
0.0970972763 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 15 |
Hb_000964_130 |
0.0980505819 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 16 |
Hb_000130_490 |
0.0983900925 |
- |
- |
PREDICTED: uncharacterized protein LOC105637408 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002007_160 |
0.0989916323 |
- |
- |
PREDICTED: uncharacterized protein LOC105647522 [Jatropha curcas] |
| 18 |
Hb_002918_180 |
0.1004179674 |
- |
- |
hypothetical protein POPTR_0004s04740g [Populus trichocarpa] |
| 19 |
Hb_000603_110 |
0.1004579999 |
- |
- |
copper transporter, putative [Ricinus communis] |
| 20 |
Hb_002063_020 |
0.1040797324 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |