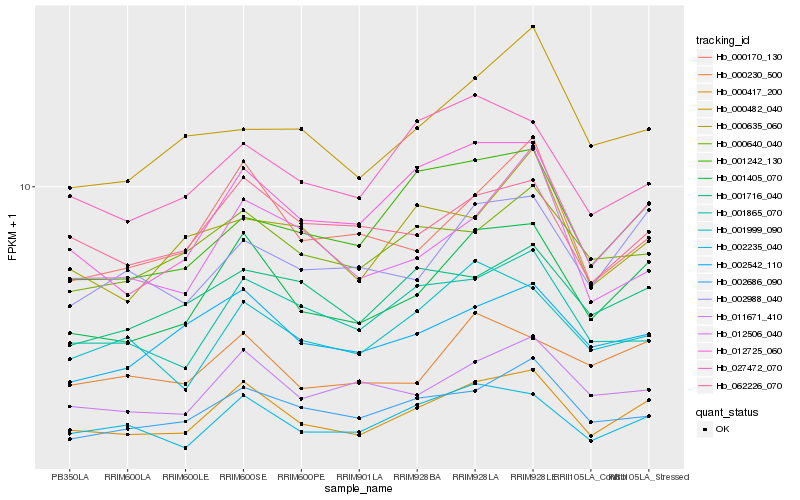
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000417_200 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001405_070 |
0.0399203102 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 3 |
Hb_012725_060 |
0.077608591 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_002235_040 |
0.0797596434 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 5 |
Hb_000170_130 |
0.0926542423 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH2 [Jatropha curcas] |
| 6 |
Hb_002686_090 |
0.0946005846 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g13650 [Jatropha curcas] |
| 7 |
Hb_001242_130 |
0.0970657614 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 4-like [Jatropha curcas] |
| 8 |
Hb_011671_410 |
0.0973888277 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 9 |
Hb_001999_090 |
0.1019752499 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001865_070 |
0.1028256856 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012506_040 |
0.1030634554 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105637346 [Jatropha curcas] |
| 12 |
Hb_002988_040 |
0.1032249207 |
- |
- |
- |
| 13 |
Hb_002542_110 |
0.1040385891 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 14 |
Hb_027472_070 |
0.1053273211 |
- |
- |
PREDICTED: uncharacterized protein LOC105646172 [Jatropha curcas] |
| 15 |
Hb_062226_070 |
0.1096911841 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000230_500 |
0.1106783607 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g19020, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000640_040 |
0.1117039912 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001716_040 |
0.1122038712 |
- |
- |
ribonuclease p/mrp subunit, putative [Ricinus communis] |
| 19 |
Hb_000635_060 |
0.1127723858 |
- |
- |
PREDICTED: uncharacterized protein LOC105644797 [Jatropha curcas] |
| 20 |
Hb_000482_040 |
0.1131788107 |
- |
- |
Protein YME1, putative [Ricinus communis] |