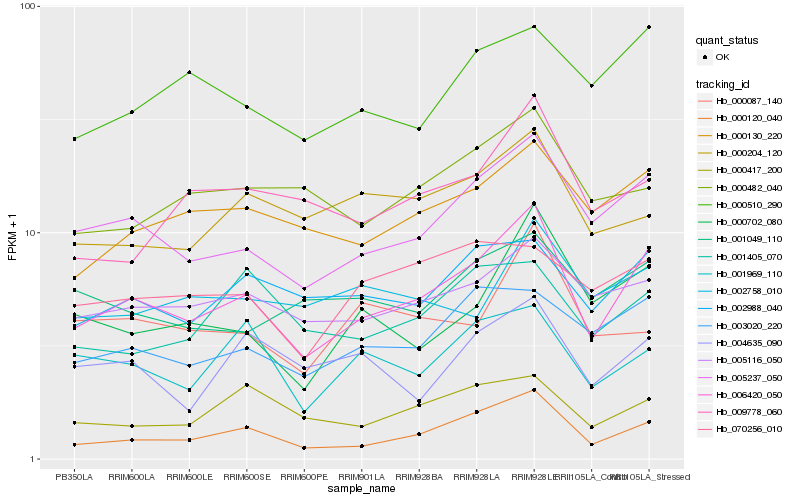
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006420_050 |
0.0 |
- |
- |
helicase, putative [Ricinus communis] |
| 2 |
Hb_005237_050 |
0.0865032042 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105630924 [Jatropha curcas] |
| 3 |
Hb_000120_040 |
0.0898361138 |
- |
- |
- |
| 4 |
Hb_000130_220 |
0.1233106225 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_003020_220 |
0.1233133053 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005116_050 |
0.1251719531 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 7 |
Hb_002988_040 |
0.1285509968 |
- |
- |
- |
| 8 |
Hb_000702_080 |
0.1344390913 |
- |
- |
PREDICTED: uncharacterized protein LOC105630492 [Jatropha curcas] |
| 9 |
Hb_000510_290 |
0.1349352467 |
- |
- |
PREDICTED: protein OSB2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_000417_200 |
0.1415199037 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: pentatricopeptide repeat-containing protein DOT4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_009778_060 |
0.142163591 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 12 |
Hb_001969_110 |
0.1431587493 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g15510, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000482_040 |
0.1448836566 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 14 |
Hb_070256_010 |
0.1468577149 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105630457 [Jatropha curcas] |
| 15 |
Hb_000204_120 |
0.1474184199 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 16 |
Hb_001049_110 |
0.1481739181 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_004635_090 |
0.1505760551 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 18 |
Hb_001405_070 |
0.1506923634 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g23020 [Jatropha curcas] |
| 19 |
Hb_002758_010 |
0.1511339517 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 20 |
Hb_000087_140 |
0.1517659173 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |