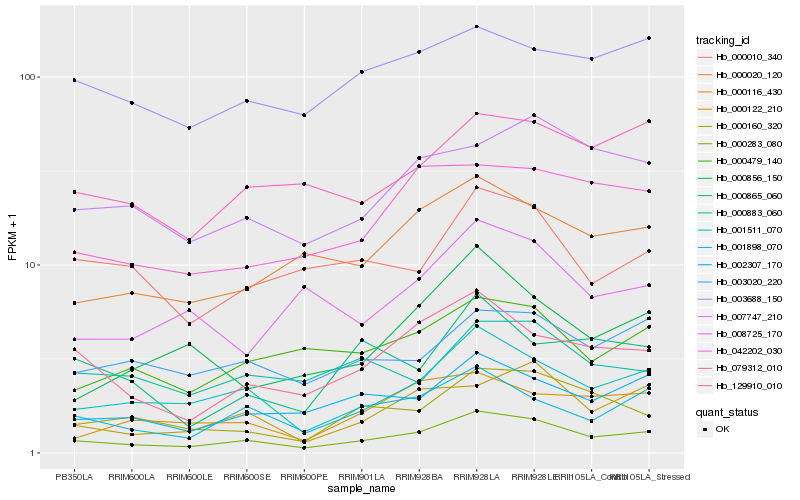
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000283_080 |
0.0 |
- |
- |
- |
| 2 |
Hb_001511_070 |
0.0967017995 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0010s19310g [Populus trichocarpa] |
| 3 |
Hb_129910_010 |
0.1234435178 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1X [Populus euphratica] |
| 4 |
Hb_002307_170 |
0.1278918054 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 5 |
Hb_001898_070 |
0.1281689658 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 6 |
Hb_000122_210 |
0.1319623372 |
- |
- |
- |
| 7 |
Hb_000020_120 |
0.1401592282 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 8, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_000160_320 |
0.1403453181 |
- |
- |
hypothetical protein POPTR_0004s08730g [Populus trichocarpa] |
| 9 |
Hb_007747_210 |
0.1457322215 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_042202_030 |
0.1465573204 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 11 |
Hb_003020_220 |
0.1468911947 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46790, chloroplastic [Jatropha curcas] |
| 12 |
Hb_079312_010 |
0.1481156136 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000479_140 |
0.1489118156 |
- |
- |
TPA: hypothetical protein ZEAMMB73_881803, partial [Zea mays] |
| 14 |
Hb_000856_150 |
0.1505516265 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 15 |
Hb_000010_340 |
0.1507067796 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000116_430 |
0.1585303419 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_003688_150 |
0.1614435861 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 18 |
Hb_000883_060 |
0.1641385294 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 19 |
Hb_000865_060 |
0.1642703413 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 20 |
Hb_008725_170 |
0.1642747111 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like [Populus euphratica] |