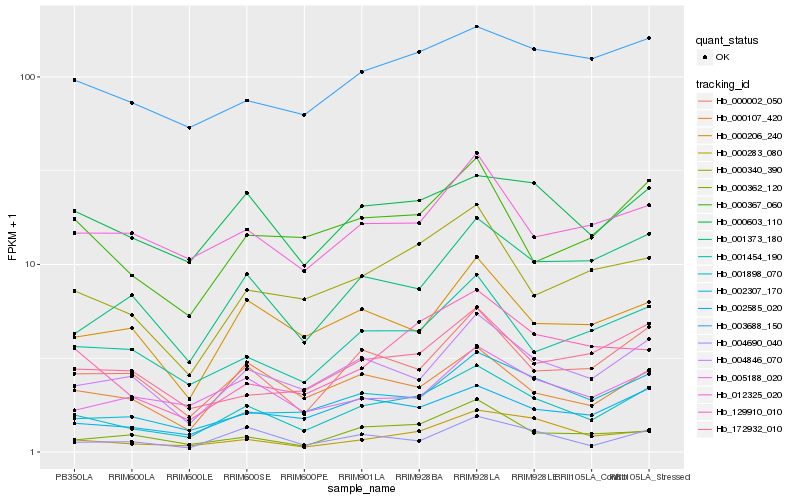
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001898_070 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g16610 [Jatropha curcas] |
| 2 |
Hb_002307_170 |
0.1073056568 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 3 |
Hb_004846_070 |
0.1082951309 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 4 |
Hb_000002_050 |
0.1104320638 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 5 |
Hb_002585_020 |
0.1116831333 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18485-like [Populus euphratica] |
| 6 |
Hb_000340_390 |
0.1180186865 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 7 |
Hb_001454_190 |
0.1221942357 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |
| 8 |
Hb_000603_110 |
0.1246283632 |
- |
- |
copper transporter, putative [Ricinus communis] |
| 9 |
Hb_000283_080 |
0.1281689658 |
- |
- |
- |
| 10 |
Hb_000367_060 |
0.128863397 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 11 |
Hb_004690_040 |
0.130070205 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 12 |
Hb_001373_180 |
0.130147625 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000107_420 |
0.1309738441 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 14 |
Hb_129910_010 |
0.1313295609 |
- |
- |
PREDICTED: F-box-like/WD repeat-containing protein TBL1X [Populus euphratica] |
| 15 |
Hb_003688_150 |
0.1314720124 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 16 |
Hb_172932_010 |
0.1343299196 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000206_240 |
0.1343453868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000362_120 |
0.1345294905 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 19 |
Hb_005188_020 |
0.1346184352 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g02330 [Jatropha curcas] |
| 20 |
Hb_012325_020 |
0.1349318288 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein At1g05910 isoform X1 [Jatropha curcas] |