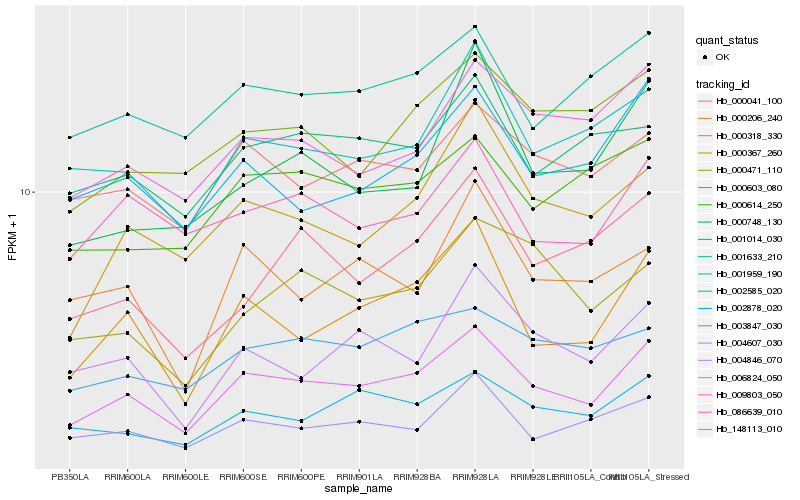
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006824_050 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000318_330 |
0.0774147082 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15630, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001959_190 |
0.0961647976 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 4 |
Hb_002585_020 |
0.0980442885 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18485-like [Populus euphratica] |
| 5 |
Hb_000367_260 |
0.0985737986 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 6 |
Hb_002878_020 |
0.1030954515 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 7 |
Hb_009803_050 |
0.1032453547 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 8 |
Hb_004607_030 |
0.1037148887 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial [Jatropha curcas] |
| 9 |
Hb_000041_100 |
0.1049479213 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 41 [Jatropha curcas] |
| 10 |
Hb_003847_030 |
0.1062444204 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001633_210 |
0.1076402121 |
- |
- |
PREDICTED: RING finger protein 10 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004846_070 |
0.1078350428 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 13 |
Hb_000603_080 |
0.108710678 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170 [Vitis vinifera] |
| 14 |
Hb_086639_010 |
0.1097615089 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000206_240 |
0.1114328039 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000614_250 |
0.1125739668 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 17 |
Hb_001014_030 |
0.1130541098 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000748_130 |
0.1140274333 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_148113_010 |
0.1140894422 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17C-like isoform X3 [Citrus sinensis] |
| 20 |
Hb_000471_110 |
0.1151594395 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39980, chloroplastic [Jatropha curcas] |