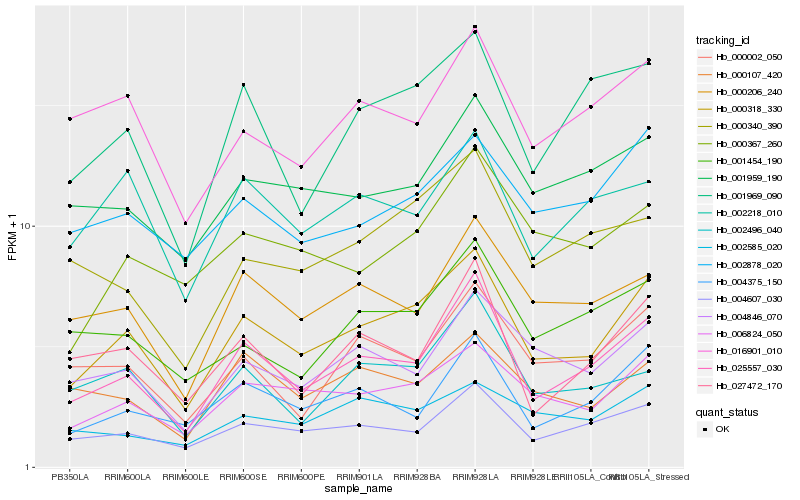
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000318_330 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15630, mitochondrial [Jatropha curcas] |
| 2 |
Hb_006824_050 |
0.0774147082 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g29760, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_025557_030 |
0.080687467 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004607_030 |
0.1048379829 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial [Jatropha curcas] |
| 5 |
Hb_004846_070 |
0.1174046544 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 6 |
Hb_001969_090 |
0.1177551552 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 7 |
Hb_002878_020 |
0.118445434 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 8 |
Hb_000206_240 |
0.1192716286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001959_190 |
0.1225206835 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 10 |
Hb_002218_010 |
0.1235971427 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 11 |
Hb_004375_150 |
0.1245423868 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 12 |
Hb_000367_260 |
0.1251838641 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 13 |
Hb_016901_010 |
0.1252179679 |
- |
- |
PREDICTED: WPP domain-interacting protein 1-like [Jatropha curcas] |
| 14 |
Hb_000002_050 |
0.1264570146 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 15 |
Hb_002585_020 |
0.1266478923 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18485-like [Populus euphratica] |
| 16 |
Hb_000107_420 |
0.127875897 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03880, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000340_390 |
0.1281492735 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 18 |
Hb_002496_040 |
0.1281952221 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 19 |
Hb_027472_170 |
0.1287518007 |
- |
- |
hypothetical protein POPTR_0007s08080g [Populus trichocarpa] |
| 20 |
Hb_001454_190 |
0.1292567697 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g06920 [Jatropha curcas] |