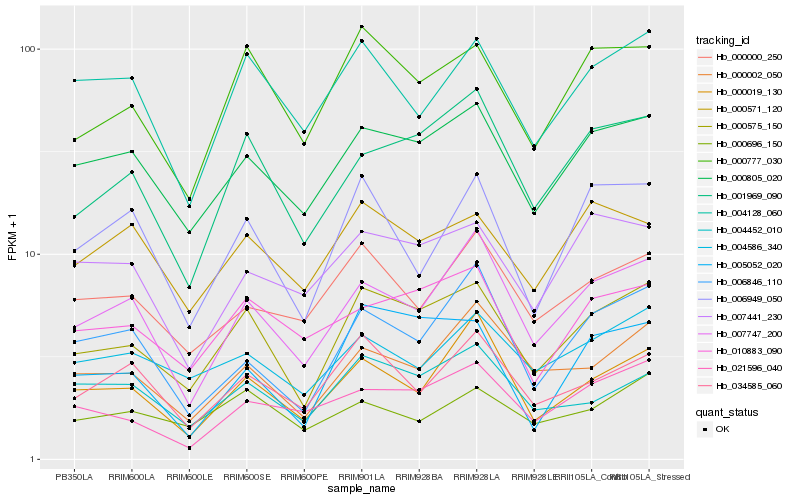
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000575_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 2 |
Hb_000777_030 |
0.0922835155 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 3 |
Hb_000805_020 |
0.0953689162 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_001969_090 |
0.1009428279 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 5 |
Hb_007747_200 |
0.1150098665 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g37570 [Jatropha curcas] |
| 6 |
Hb_004452_010 |
0.1201568121 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 7 |
Hb_010883_090 |
0.1216292609 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000002_050 |
0.1224600559 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 9 |
Hb_004128_060 |
0.1240680341 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit A [Jatropha curcas] |
| 10 |
Hb_004586_340 |
0.1253622413 |
- |
- |
PREDICTED: serine/threonine-protein kinase PEPKR2 [Jatropha curcas] |
| 11 |
Hb_005052_020 |
0.1256738098 |
- |
- |
- |
| 12 |
Hb_000571_120 |
0.1303634146 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 13 |
Hb_034585_060 |
0.1314191522 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g50420 [Jatropha curcas] |
| 14 |
Hb_000696_150 |
0.1316658268 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g51320 [Jatropha curcas] |
| 15 |
Hb_000019_130 |
0.1319886669 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At2g01510 [Jatropha curcas] |
| 16 |
Hb_006949_050 |
0.1321216756 |
- |
- |
unknown [Lotus japonicus] |
| 17 |
Hb_000000_250 |
0.1335327131 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g19290 [Jatropha curcas] |
| 18 |
Hb_021596_040 |
0.1344829932 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_006846_110 |
0.1349184075 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61800 [Jatropha curcas] |
| 20 |
Hb_007441_230 |
0.1349401759 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase [Jatropha curcas] |