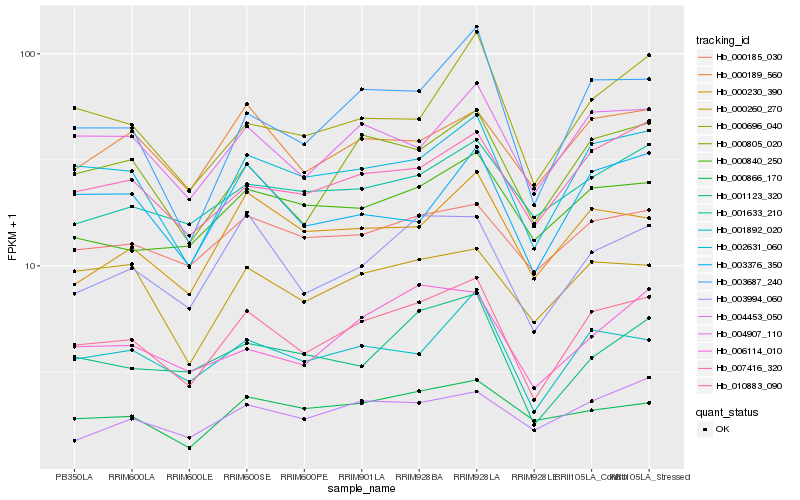
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002631_060 |
0.0639310144 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 3 |
Hb_003994_060 |
0.0750315134 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 4 |
Hb_000805_020 |
0.0767769041 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_003687_240 |
0.082049264 |
- |
- |
PREDICTED: cryptochrome-1 [Jatropha curcas] |
| 6 |
Hb_001892_020 |
0.0860215631 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_004907_110 |
0.0863294126 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007416_320 |
0.0879093832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006114_010 |
0.0951735549 |
- |
- |
PREDICTED: uncharacterized protein LOC105629068 [Jatropha curcas] |
| 10 |
Hb_001123_320 |
0.0958884341 |
- |
- |
PREDICTED: uncharacterized protein LOC105647314 isoform X3 [Jatropha curcas] |
| 11 |
Hb_000189_560 |
0.0979631239 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1 [Jatropha curcas] |
| 12 |
Hb_000230_390 |
0.0980034282 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 13 |
Hb_000840_250 |
0.0982269609 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 14 |
Hb_000260_270 |
0.0985238205 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 15 |
Hb_000185_030 |
0.0988436268 |
- |
- |
PREDICTED: arginine-glutamic acid dipeptide repeats protein-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001633_210 |
0.1009304338 |
- |
- |
PREDICTED: RING finger protein 10 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003376_350 |
0.1015781282 |
- |
- |
PREDICTED: CASP-like protein 5A1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000696_040 |
0.1019722294 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_004453_050 |
0.1026248506 |
- |
- |
PREDICTED: uncharacterized protein LOC105637675 [Jatropha curcas] |
| 20 |
Hb_000866_170 |
0.1044583814 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |