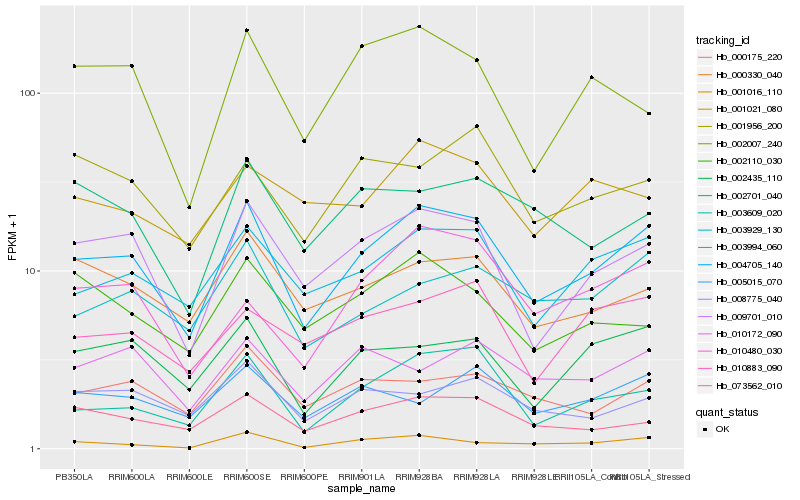
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004705_140 |
0.0 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 2 |
Hb_009701_010 |
0.1007011542 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 3 |
Hb_003994_060 |
0.1059013065 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 4 |
Hb_003609_020 |
0.1217654144 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 5 |
Hb_073562_010 |
0.1244720331 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 6 |
Hb_002435_110 |
0.1270033768 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 7 |
Hb_003929_130 |
0.1317306737 |
- |
- |
PREDICTED: uncharacterized protein LOC105643628 [Jatropha curcas] |
| 8 |
Hb_000330_040 |
0.13213368 |
- |
- |
PREDICTED: protein RER1B-like [Jatropha curcas] |
| 9 |
Hb_000175_220 |
0.1322192074 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 10 |
Hb_001016_110 |
0.1375991099 |
desease resistance |
Gene Name: Lectin_legB |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 11 |
Hb_008775_040 |
0.138031041 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 12 |
Hb_010883_090 |
0.1401119307 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005015_070 |
0.1433640969 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 14 |
Hb_002007_240 |
0.1438095466 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 15 |
Hb_001956_200 |
0.1444294419 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_010172_090 |
0.1452509214 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_002701_040 |
0.1459041044 |
- |
- |
PREDICTED: auxin transport protein BIG [Jatropha curcas] |
| 18 |
Hb_010480_030 |
0.1464952577 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_001021_080 |
0.1470476845 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 20 |
Hb_002110_030 |
0.1472269357 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |