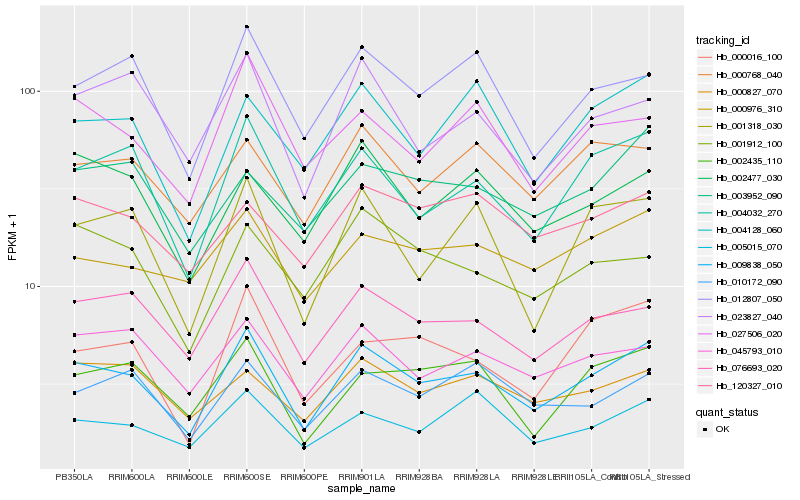
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009838_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004032_270 |
0.0860847592 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_076693_020 |
0.0937156804 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 4 |
Hb_012807_050 |
0.1031194312 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 5 |
Hb_001318_030 |
0.1038397671 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 6 |
Hb_045793_010 |
0.1077916818 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Pyrus x bretschneideri] |
| 7 |
Hb_002435_110 |
0.1087971621 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 8 |
Hb_023827_040 |
0.1103400413 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 9 |
Hb_027506_020 |
0.1107098262 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 10 |
Hb_005015_070 |
0.1113001939 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g39530 [Jatropha curcas] |
| 11 |
Hb_004128_060 |
0.1121801157 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit A [Jatropha curcas] |
| 12 |
Hb_000016_100 |
0.112673189 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP19 [Jatropha curcas] |
| 13 |
Hb_010172_090 |
0.1137446573 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000976_310 |
0.1138114313 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000768_040 |
0.1141878545 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 16 |
Hb_002477_030 |
0.1145403628 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105631404 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001912_100 |
0.116349833 |
- |
- |
PREDICTED: uncharacterized protein LOC105636609 [Jatropha curcas] |
| 18 |
Hb_000827_070 |
0.1188367789 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 19 |
Hb_120327_010 |
0.1215675912 |
- |
- |
PREDICTED: uncharacterized protein LOC105650959 [Jatropha curcas] |
| 20 |
Hb_003952_090 |
0.1216729038 |
- |
- |
PREDICTED: threonine dehydratase biosynthetic, chloroplastic [Jatropha curcas] |