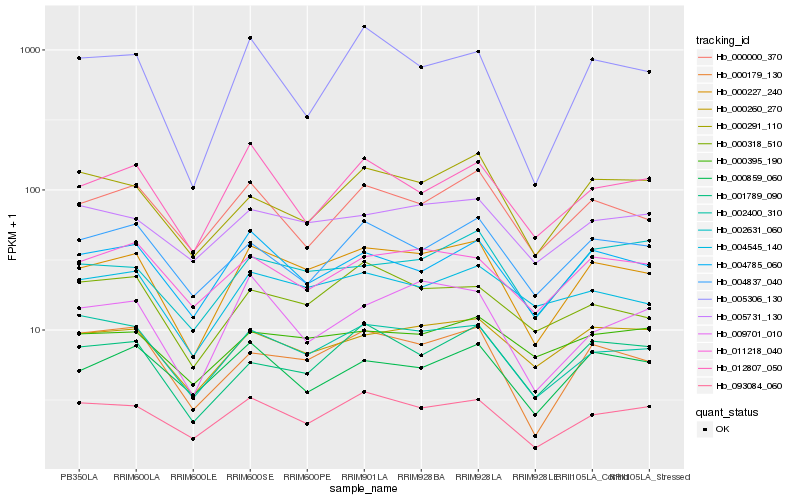
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000227_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_02139 [Jatropha curcas] |
| 2 |
Hb_093084_060 |
0.0881564474 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 3 |
Hb_004785_060 |
0.0935075538 |
- |
- |
PREDICTED: SKP1-interacting partner 15 [Jatropha curcas] |
| 4 |
Hb_002400_310 |
0.0939174288 |
- |
- |
hypothetical protein JCGZ_07138 [Jatropha curcas] |
| 5 |
Hb_000179_130 |
0.0941798001 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 6 |
Hb_000260_270 |
0.1009069102 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_004545_140 |
0.1027007415 |
- |
- |
hypothetical protein JCGZ_23405 [Jatropha curcas] |
| 8 |
Hb_000000_370 |
0.1073861789 |
transcription factor |
TF Family: BES1 |
PREDICTED: protein BRASSINAZOLE-RESISTANT 1 [Jatropha curcas] |
| 9 |
Hb_005731_130 |
0.1079476095 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL6 [Jatropha curcas] |
| 10 |
Hb_000318_510 |
0.1097631746 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 11 |
Hb_001789_090 |
0.1103506947 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105648450 [Jatropha curcas] |
| 12 |
Hb_011218_040 |
0.1106122005 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 13 |
Hb_009701_010 |
0.111332949 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 14 |
Hb_004837_040 |
0.1126735376 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 1-like [Prunus mume] |
| 15 |
Hb_000859_060 |
0.113143526 |
- |
- |
PREDICTED: uncharacterized protein LOC105641312 [Jatropha curcas] |
| 16 |
Hb_012807_050 |
0.1133164919 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 17 |
Hb_002631_060 |
0.1135079595 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 18 |
Hb_000291_110 |
0.1137378812 |
- |
- |
eukaryotic translation initiation factor [Hevea brasiliensis] |
| 19 |
Hb_005306_130 |
0.1155723685 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000395_190 |
0.1160957959 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 7-like [Jatropha curcas] |