| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
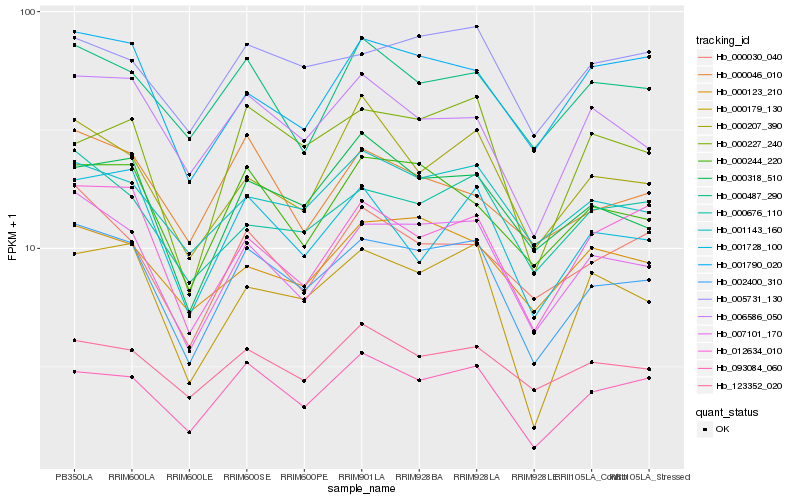
Hb_002400_310 |
0.0 |
- |
- |
hypothetical protein JCGZ_07138 [Jatropha curcas] |
| 2 |
Hb_007101_170 |
0.0537868932 |
- |
- |
hypothetical protein POPTR_0010s10490g [Populus trichocarpa] |
| 3 |
Hb_093084_060 |
0.070487841 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 4 |
Hb_000244_220 |
0.0807434251 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_000318_510 |
0.0833999713 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 6 |
Hb_006586_050 |
0.0860087064 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 7 |
Hb_001143_160 |
0.0861117276 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000676_110 |
0.0861429904 |
- |
- |
glycosyl transferase family 1 family protein [Populus trichocarpa] |
| 9 |
Hb_000179_130 |
0.0868910678 |
- |
- |
PREDICTED: protein Brevis radix-like 4 [Jatropha curcas] |
| 10 |
Hb_001728_100 |
0.0902532998 |
- |
- |
hypothetical protein JCGZ_03429 [Jatropha curcas] |
| 11 |
Hb_005731_130 |
0.0908682379 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL6 [Jatropha curcas] |
| 12 |
Hb_012634_010 |
0.0910502653 |
- |
- |
PREDICTED: uncharacterized protein LOC105110292 [Populus euphratica] |
| 13 |
Hb_000030_040 |
0.0913899902 |
- |
- |
yth domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000227_240 |
0.0939174288 |
- |
- |
hypothetical protein JCGZ_02139 [Jatropha curcas] |
| 15 |
Hb_001790_020 |
0.09442326 |
- |
- |
PREDICTED: uncharacterized protein LOC105642165 [Jatropha curcas] |
| 16 |
Hb_000207_390 |
0.0960788125 |
- |
- |
PREDICTED: uncharacterized protein LOC105637143 [Jatropha curcas] |
| 17 |
Hb_123352_020 |
0.096798941 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560 [Jatropha curcas] |
| 18 |
Hb_000123_210 |
0.09790506 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000487_290 |
0.0984848321 |
- |
- |
ribonucleic acid binding protein S1, putative [Ricinus communis] |
| 20 |
Hb_000046_010 |
0.099479822 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |