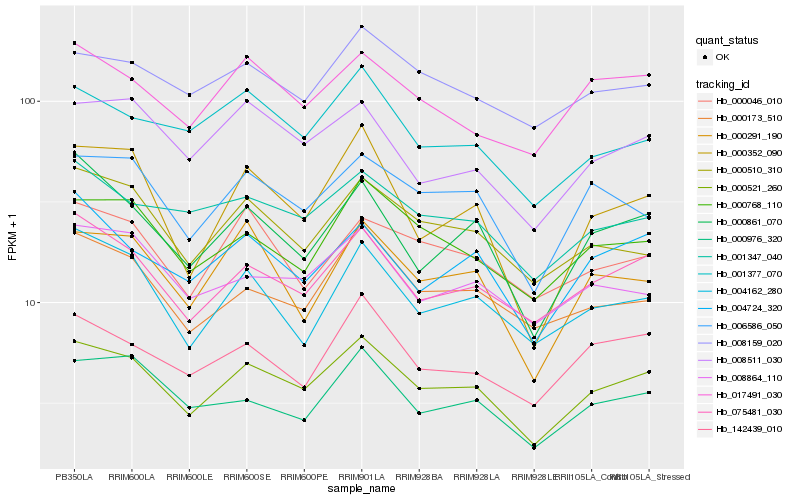
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000521_260 |
0.0 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_075481_030 |
0.0655611069 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000768_110 |
0.0743767488 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006586_050 |
0.0748473821 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 5 |
Hb_000510_310 |
0.0795469716 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 6 |
Hb_008511_030 |
0.0811543498 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 7 |
Hb_000976_320 |
0.0818255527 |
- |
- |
UPF0061 protein azo1574 [Morus notabilis] |
| 8 |
Hb_017491_030 |
0.0819398354 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 9 |
Hb_000173_510 |
0.0824241835 |
- |
- |
PREDICTED: AP-5 complex subunit mu [Jatropha curcas] |
| 10 |
Hb_001347_040 |
0.0828442875 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 11 |
Hb_000861_070 |
0.0834914054 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_142439_010 |
0.083782132 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 13 |
Hb_001377_070 |
0.0847874091 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004724_320 |
0.0849129318 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 15 |
Hb_008864_110 |
0.0861720657 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 16 |
Hb_004162_280 |
0.0867271494 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008159_020 |
0.0868450504 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 18 |
Hb_000291_190 |
0.0879821554 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 19 |
Hb_000352_090 |
0.0886072796 |
- |
- |
PREDICTED: syntaxin-22-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000046_010 |
0.0888754972 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1 [Jatropha curcas] |