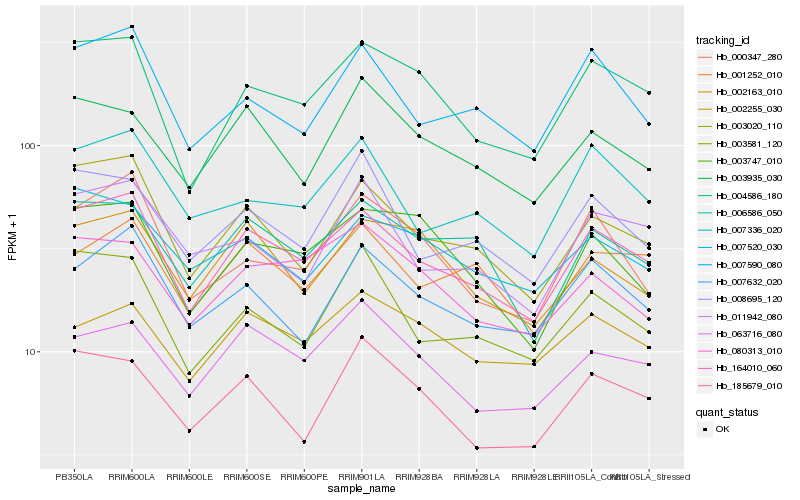
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164010_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633115 [Jatropha curcas] |
| 2 |
Hb_003020_110 |
0.0650119119 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 3 |
Hb_004586_180 |
0.0652109587 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 4 |
Hb_006586_050 |
0.0773336519 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 5 |
Hb_007590_080 |
0.0799778583 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-4 [Malus domestica] |
| 6 |
Hb_007632_020 |
0.0802385996 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 7 |
Hb_063716_080 |
0.0820948336 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 8 |
Hb_007336_020 |
0.0837444368 |
- |
- |
Mannose-P-dolichol utilization defect 1 protein, putative [Ricinus communis] |
| 9 |
Hb_003581_120 |
0.0846005653 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 10 |
Hb_001252_010 |
0.0853518189 |
- |
- |
hypothetical protein JCGZ_01389 [Jatropha curcas] |
| 11 |
Hb_008695_120 |
0.0866888504 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_080313_010 |
0.0883370045 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 13 |
Hb_007520_030 |
0.0883373468 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 14 |
Hb_003747_010 |
0.0901398199 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 15 |
Hb_002163_010 |
0.0908277418 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 16 |
Hb_003935_030 |
0.0911911832 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 17 |
Hb_002255_030 |
0.0912196578 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 18 |
Hb_000347_280 |
0.0914938534 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 19 |
Hb_011942_080 |
0.0918113575 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 20 |
Hb_185679_010 |
0.0918616887 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |