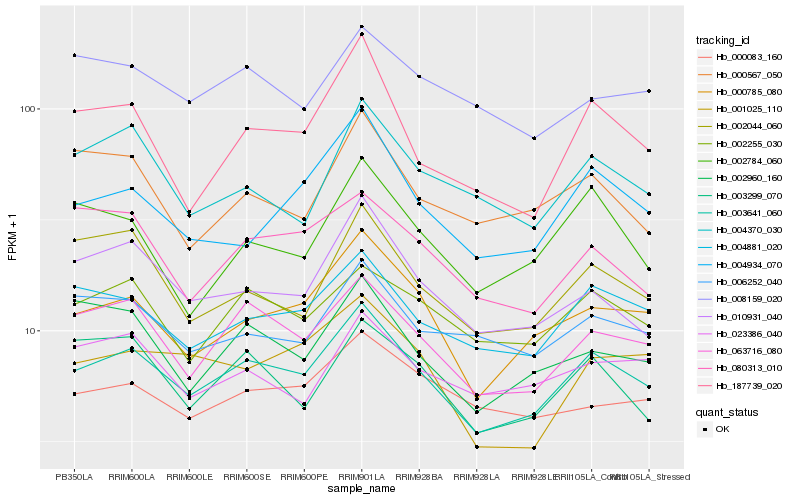
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003641_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 2 |
Hb_063716_080 |
0.0661515071 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 3 |
Hb_000785_080 |
0.0722960788 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 4 |
Hb_004881_020 |
0.073993281 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 5 |
Hb_002255_030 |
0.0805722927 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 6 |
Hb_010931_040 |
0.0867508225 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 7 |
Hb_187739_020 |
0.0910284482 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 8 |
Hb_004934_070 |
0.093035358 |
- |
- |
PREDICTED: U2 small nuclear ribonucleoprotein A' [Jatropha curcas] |
| 9 |
Hb_002044_060 |
0.093099624 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 10 |
Hb_002960_160 |
0.0933767478 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 11 |
Hb_006252_040 |
0.0939386521 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_003299_070 |
0.0951917133 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 13 |
Hb_000083_160 |
0.0952878457 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 14 |
Hb_004370_030 |
0.0966706714 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 15 |
Hb_023386_040 |
0.0979490216 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 16 |
Hb_080313_010 |
0.0979751797 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 17 |
Hb_001025_110 |
0.0987887669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002784_060 |
0.0988664855 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008159_020 |
0.0993186485 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 20 |
Hb_000567_050 |
0.0997349246 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |