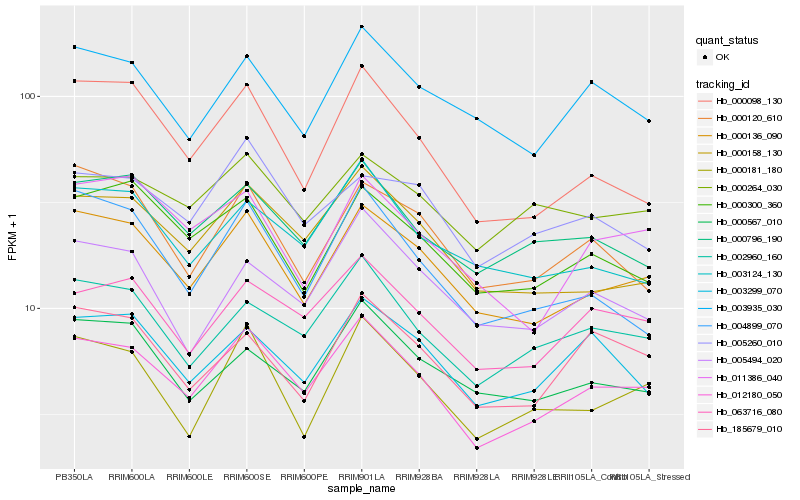
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012180_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632355 isoform X3 [Jatropha curcas] |
| 2 |
Hb_000136_090 |
0.0655654462 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 3 |
Hb_063716_080 |
0.0753930557 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 4 |
Hb_002960_160 |
0.0782309645 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 5 |
Hb_000158_130 |
0.081262178 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000098_130 |
0.0860891459 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 7 |
Hb_000796_190 |
0.0884993818 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 8 |
Hb_000181_180 |
0.0922986388 |
- |
- |
PREDICTED: uncharacterized protein LOC105637608 [Jatropha curcas] |
| 9 |
Hb_003124_130 |
0.0956971807 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003299_070 |
0.0958522181 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 11 |
Hb_003935_030 |
0.0964848889 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 12 |
Hb_000300_360 |
0.096823685 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 13 |
Hb_185679_010 |
0.0991798109 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |
| 14 |
Hb_004899_070 |
0.0993959928 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 15 |
Hb_011386_040 |
0.0995161621 |
- |
- |
PREDICTED: uncharacterized protein LOC105639243 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000120_610 |
0.1007229446 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 17 |
Hb_005494_020 |
0.1016087062 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 18 |
Hb_000567_010 |
0.1016358915 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 19 |
Hb_005260_010 |
0.1019318974 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000264_030 |
0.1034653927 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |