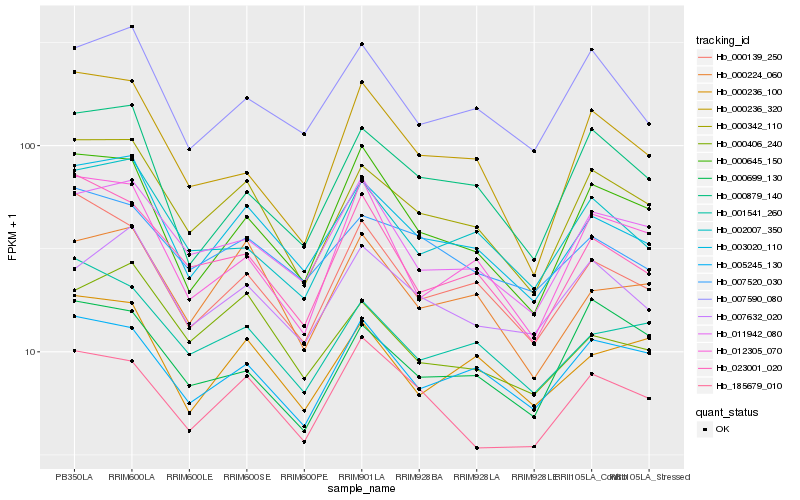
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000342_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638174 [Jatropha curcas] |
| 2 |
Hb_003020_110 |
0.0653301094 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 3 |
Hb_002007_350 |
0.0809745648 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 1-like [Jatropha curcas] |
| 4 |
Hb_005245_130 |
0.0829203982 |
- |
- |
PREDICTED: uncharacterized protein LOC105643730 [Jatropha curcas] |
| 5 |
Hb_001541_260 |
0.082962786 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 6 |
Hb_011942_080 |
0.0851143474 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 7 |
Hb_000139_250 |
0.0861643143 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 8 |
Hb_023001_020 |
0.08638549 |
- |
- |
PREDICTED: mitochondrial succinate-fumarate transporter 1 [Jatropha curcas] |
| 9 |
Hb_000224_060 |
0.0881487654 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 10 |
Hb_000879_140 |
0.0884109459 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0283697-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_000645_150 |
0.0884720247 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 12 |
Hb_000699_130 |
0.0896316128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000236_320 |
0.0908071393 |
- |
- |
PREDICTED: ER membrane protein complex subunit 8/9 homolog [Jatropha curcas] |
| 14 |
Hb_000236_100 |
0.0936448321 |
- |
- |
Dynein alpha chain, flagellar outer arm [Gossypium arboreum] |
| 15 |
Hb_007590_080 |
0.0939444764 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-4 [Malus domestica] |
| 16 |
Hb_007520_030 |
0.094135954 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 17 |
Hb_007632_020 |
0.0947738764 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 18 |
Hb_000406_240 |
0.0949066584 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2-like [Jatropha curcas] |
| 19 |
Hb_012305_070 |
0.0961563908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_185679_010 |
0.0963391219 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |