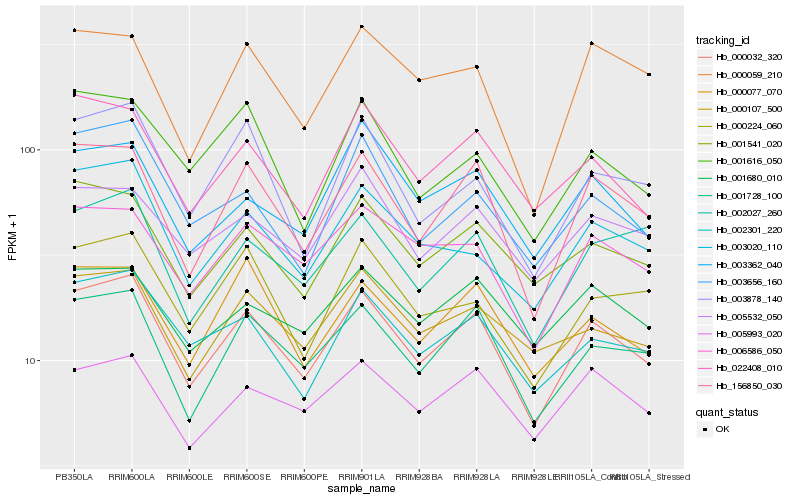
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000032_320 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_156850_030 |
0.0509322811 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 3 |
Hb_001728_100 |
0.0604011843 |
- |
- |
hypothetical protein JCGZ_03429 [Jatropha curcas] |
| 4 |
Hb_002301_220 |
0.0731075847 |
transcription factor |
TF Family: C3H |
PREDICTED: protein FRIGIDA-ESSENTIAL 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001541_020 |
0.074557352 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 6 |
Hb_022408_010 |
0.075840052 |
- |
- |
PREDICTED: protein SUPPRESSOR OF GENE SILENCING 3 [Jatropha curcas] |
| 7 |
Hb_006586_050 |
0.0770416071 |
- |
- |
PREDICTED: probable protein phosphatase 2C 39 [Jatropha curcas] |
| 8 |
Hb_000077_070 |
0.0776455308 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003020_110 |
0.0799668526 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 10 |
Hb_005993_020 |
0.0821079894 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 11 |
Hb_000107_500 |
0.082324495 |
- |
- |
PREDICTED: protein FAR1-RELATED SEQUENCE 9 [Jatropha curcas] |
| 12 |
Hb_005532_050 |
0.0826742275 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_003878_140 |
0.0836613516 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like F-box protein 7 [Jatropha curcas] |
| 14 |
Hb_000224_060 |
0.0839058854 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6 [Jatropha curcas] |
| 15 |
Hb_003362_040 |
0.0843412403 |
- |
- |
PREDICTED: probable methionine--tRNA ligase [Jatropha curcas] |
| 16 |
Hb_002027_260 |
0.084395963 |
- |
- |
PREDICTED: gibberellin receptor GID1B [Jatropha curcas] |
| 17 |
Hb_000059_210 |
0.084953189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001680_010 |
0.0855527399 |
- |
- |
Serine/threonine protein kinase SAPK3 [Medicago truncatula] |
| 19 |
Hb_003656_160 |
0.0855808713 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein B456_013G226400 [Gossypium raimondii] |
| 20 |
Hb_001616_050 |
0.087485107 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |