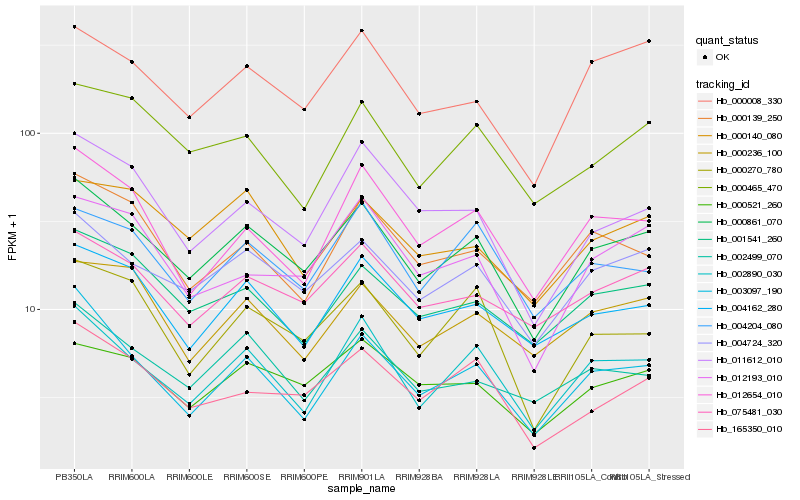
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000861_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_004724_320 |
0.0500093152 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 3 |
Hb_002890_030 |
0.0752397499 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RGA2, putative [Ricinus communis] |
| 4 |
Hb_075481_030 |
0.0768961168 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000521_260 |
0.0834914054 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000008_330 |
0.0859688954 |
- |
- |
60S ribosomal protein L12A [Hevea brasiliensis] |
| 7 |
Hb_004162_280 |
0.0864601378 |
- |
- |
PREDICTED: uncharacterized protein LOC105628004 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001541_260 |
0.0876514293 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 9 |
Hb_165350_010 |
0.0926649782 |
- |
- |
unnamed protein product [Triticum aestivum] |
| 10 |
Hb_002499_070 |
0.0927024215 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 11 |
Hb_012654_010 |
0.0965871037 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 12 |
Hb_000465_470 |
0.0966002387 |
- |
- |
unknown [Picea sitchensis] |
| 13 |
Hb_004204_080 |
0.0968643119 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 14 |
Hb_000236_100 |
0.0970086701 |
- |
- |
Dynein alpha chain, flagellar outer arm [Gossypium arboreum] |
| 15 |
Hb_003097_190 |
0.0971006455 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 16 |
Hb_000270_780 |
0.0972416927 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 17 |
Hb_011612_010 |
0.0988709855 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 18 |
Hb_000140_080 |
0.0990172232 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_012193_010 |
0.0996558539 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000139_250 |
0.100112373 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |