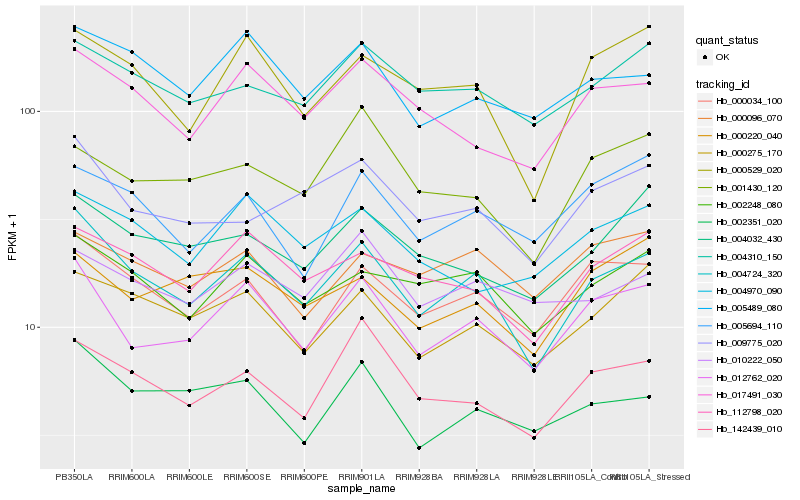
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012762_020 |
0.0 |
- |
- |
(Di)nucleoside polyphosphate hydrolase, putative [Ricinus communis] |
| 2 |
Hb_004724_320 |
0.0763099872 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 3 |
Hb_000034_100 |
0.0871056343 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 4 |
Hb_000220_040 |
0.0976933705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_017491_030 |
0.0988793152 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 6 |
Hb_000275_170 |
0.1004407684 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b-like [Jatropha curcas] |
| 7 |
Hb_002248_080 |
0.1005653663 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 25 isoform X2 [Jatropha curcas] |
| 8 |
Hb_005694_110 |
0.1006714352 |
- |
- |
hypothetical protein PRUPE_ppa008137mg [Prunus persica] |
| 9 |
Hb_142439_010 |
0.1017031227 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 10 |
Hb_112798_020 |
0.1021242296 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 11 |
Hb_000529_020 |
0.1035421649 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 12 |
Hb_002351_020 |
0.1040107582 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |
| 13 |
Hb_000096_070 |
0.1046132013 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 14 |
Hb_004310_150 |
0.1049798794 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 15 |
Hb_005489_080 |
0.1055801051 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004970_090 |
0.1069703316 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 17 |
Hb_001430_120 |
0.1071157615 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 18 |
Hb_004032_430 |
0.1081108281 |
- |
- |
PREDICTED: uncharacterized protein LOC105635169 [Jatropha curcas] |
| 19 |
Hb_009775_020 |
0.1081316811 |
- |
- |
PREDICTED: vesicle transport protein GOT1B-like [Jatropha curcas] |
| 20 |
Hb_010222_050 |
0.1087917364 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X2 [Jatropha curcas] |