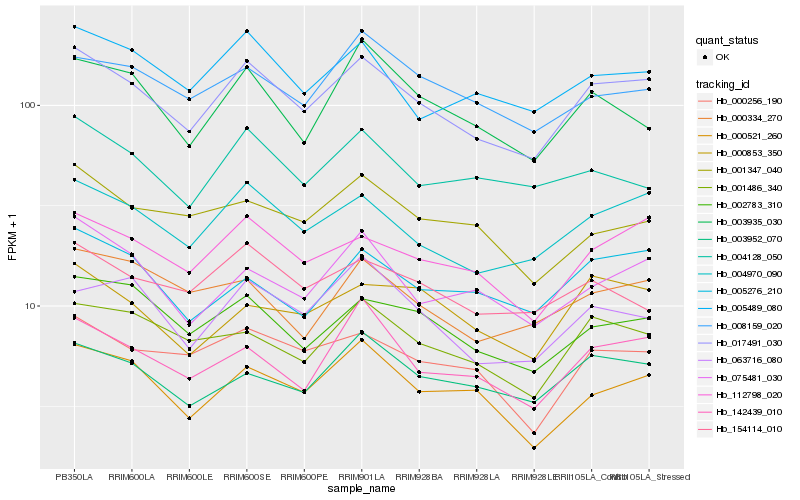
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017491_030 |
0.0 |
- |
- |
PREDICTED: 40S ribosomal protein SA-like [Jatropha curcas] |
| 2 |
Hb_004970_090 |
0.0514255285 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 3 |
Hb_112798_020 |
0.0638633579 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 4 |
Hb_002783_310 |
0.0754456506 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 5 |
Hb_005489_080 |
0.0765800205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003952_070 |
0.0780054776 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_001486_340 |
0.0786620721 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 8 |
Hb_142439_010 |
0.0809500555 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 9 |
Hb_000256_190 |
0.0809990334 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g66345, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000521_260 |
0.0819398354 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_075481_030 |
0.0829373061 |
- |
- |
PREDICTED: DNA-binding protein RHL1 isoform X3 [Jatropha curcas] |
| 12 |
Hb_005276_210 |
0.0854399159 |
- |
- |
PREDICTED: DNA repair helicase UVH6 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000853_350 |
0.086469191 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like [Jatropha curcas] |
| 14 |
Hb_008159_020 |
0.0869629889 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 15 |
Hb_063716_080 |
0.0871718467 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 16 |
Hb_004128_050 |
0.0879208019 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_154114_010 |
0.0883141756 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 18 |
Hb_000334_270 |
0.0886144853 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001347_040 |
0.0901109025 |
- |
- |
PREDICTED: uncharacterized protein LOC105633920 [Jatropha curcas] |
| 20 |
Hb_003935_030 |
0.0904261792 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |