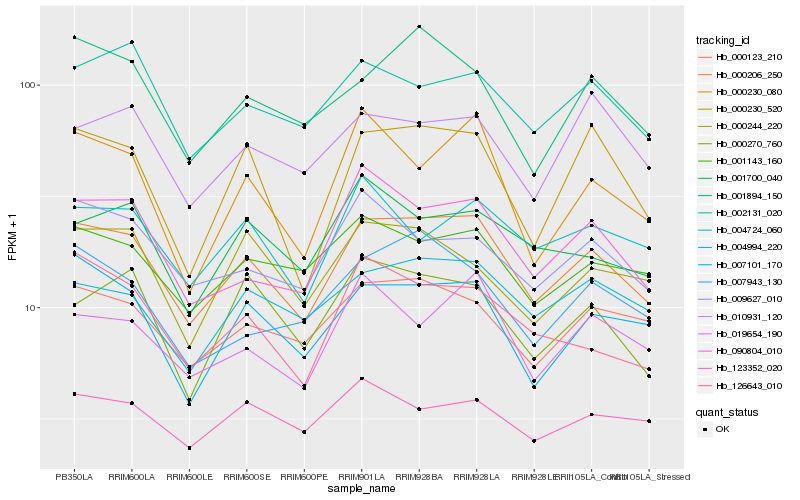
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000206_250 |
0.0 |
- |
- |
PREDICTED: UBP1-associated protein 2A-like [Jatropha curcas] |
| 2 |
Hb_000123_210 |
0.0807838198 |
- |
- |
PREDICTED: uncharacterized protein LOC105641576 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007101_170 |
0.08470968 |
- |
- |
hypothetical protein POPTR_0010s10490g [Populus trichocarpa] |
| 4 |
Hb_090804_010 |
0.0847164156 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_004994_220 |
0.0850265738 |
- |
- |
PREDICTED: K(+) efflux antiporter 2, chloroplastic-like isoform X2 [Jatropha curcas] |
| 6 |
Hb_001143_160 |
0.0856932528 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002131_020 |
0.0877529446 |
transcription factor |
TF Family: IWS1 |
PREDICTED: protein IWS1 homolog [Jatropha curcas] |
| 8 |
Hb_000230_520 |
0.0905442844 |
- |
- |
PREDICTED: serine incorporator 3 [Jatropha curcas] |
| 9 |
Hb_001894_150 |
0.0911062414 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 10 |
Hb_019654_190 |
0.0940359205 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 11 |
Hb_009627_010 |
0.0952344906 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 12 |
Hb_007943_130 |
0.0953055349 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 13 |
Hb_123352_020 |
0.096595011 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g65560 [Jatropha curcas] |
| 14 |
Hb_004724_060 |
0.0970581453 |
- |
- |
PREDICTED: uncharacterized protein LOC105640155 isoform X1 [Jatropha curcas] |
| 15 |
Hb_126643_010 |
0.0973991509 |
- |
- |
Protein transport protein Sec24C, putative [Ricinus communis] |
| 16 |
Hb_000270_760 |
0.097653886 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001700_040 |
0.0982751536 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 18 |
Hb_000230_080 |
0.0983223676 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 19 |
Hb_000244_220 |
0.0988343494 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_010931_120 |
0.0992053713 |
- |
- |
myosin heavy chain-related family protein [Populus trichocarpa] |