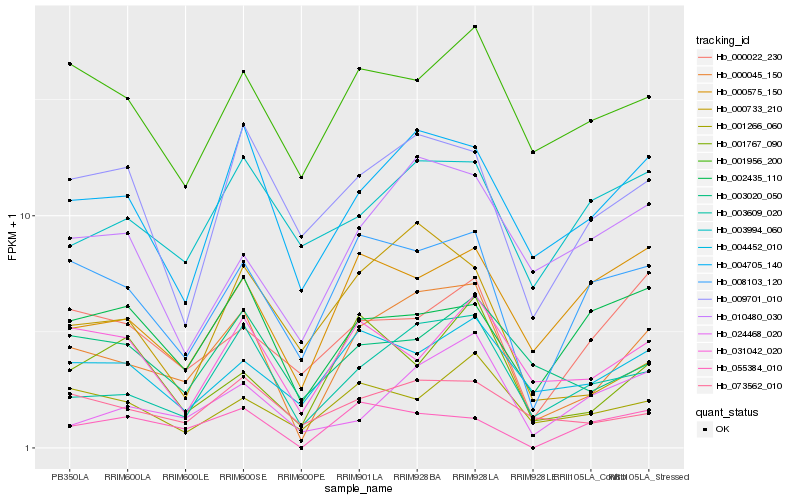
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000045_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 2 |
Hb_003609_020 |
0.1345923695 |
desease resistance |
Gene Name: Clusterin |
hypothetical protein CISIN_1g001897mg [Citrus sinensis] |
| 3 |
Hb_004705_140 |
0.1631468102 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105647306 [Jatropha curcas] |
| 4 |
Hb_008103_120 |
0.1727386007 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 5 |
Hb_073562_010 |
0.1816540044 |
- |
- |
hypothetical protein POPTR_0011s036602g, partial [Populus trichocarpa] |
| 6 |
Hb_004452_010 |
0.1936647641 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 7 |
Hb_001266_060 |
0.1942092296 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 8 |
Hb_001956_200 |
0.1955854109 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_009701_010 |
0.1971953086 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 10 |
Hb_003020_050 |
0.1978536422 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000575_150 |
0.2002282253 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 12 |
Hb_010480_030 |
0.20027528 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |
| 13 |
Hb_031042_020 |
0.2019609483 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 14 |
Hb_000733_210 |
0.2070776496 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 15 |
Hb_000022_230 |
0.2079643638 |
- |
- |
hypothetical protein RCOM_0803470 [Ricinus communis] |
| 16 |
Hb_024468_020 |
0.2092757361 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 17 |
Hb_002435_110 |
0.2100822601 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 18 |
Hb_003994_060 |
0.2101892795 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 19 |
Hb_001767_090 |
0.2104802845 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_055384_010 |
0.2120077356 |
- |
- |
- |