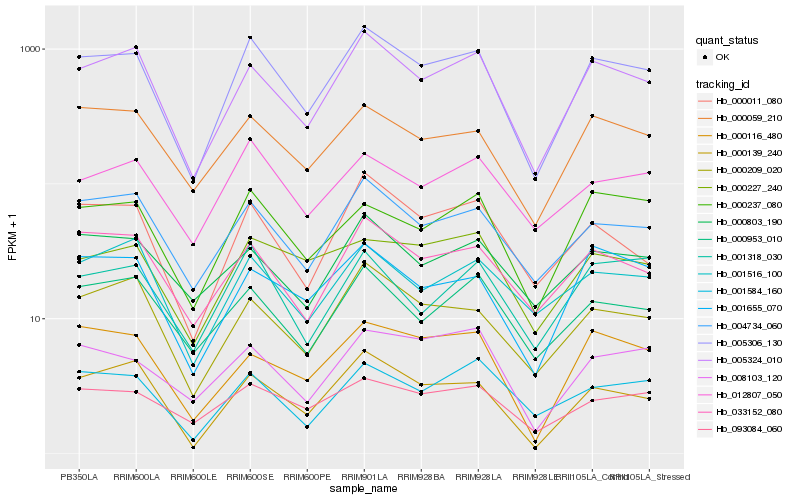
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_130 |
0.0 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 2 |
Hb_005324_010 |
0.0710334174 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 3 |
Hb_004734_060 |
0.0884725046 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 4 |
Hb_033152_080 |
0.0991606603 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 5 |
Hb_000209_020 |
0.0992567935 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000139_240 |
0.1001695887 |
- |
- |
PREDICTED: putative F-box protein At3g16210 [Jatropha curcas] |
| 7 |
Hb_000059_210 |
0.1005095306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000237_080 |
0.1041153964 |
transcription factor |
TF Family: PHD |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 9 |
Hb_000011_080 |
0.1051306021 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 10 |
Hb_093084_060 |
0.1058058743 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At1g64310 [Jatropha curcas] |
| 11 |
Hb_001655_070 |
0.1080647414 |
- |
- |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 12 |
Hb_012807_050 |
0.1093792435 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 13 |
Hb_000953_010 |
0.1111617585 |
- |
- |
hypothetical protein CISIN_1g0425012mg, partial [Citrus sinensis] |
| 14 |
Hb_001584_160 |
0.111807542 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g56570 [Jatropha curcas] |
| 15 |
Hb_001516_100 |
0.1151010123 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000116_480 |
0.1155474966 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 17 |
Hb_000227_240 |
0.1155723685 |
- |
- |
hypothetical protein JCGZ_02139 [Jatropha curcas] |
| 18 |
Hb_008103_120 |
0.1156859791 |
- |
- |
hypothetical protein CISIN_1g009650mg [Citrus sinensis] |
| 19 |
Hb_001318_030 |
0.1178890248 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 20 |
Hb_000803_190 |
0.1182475874 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |