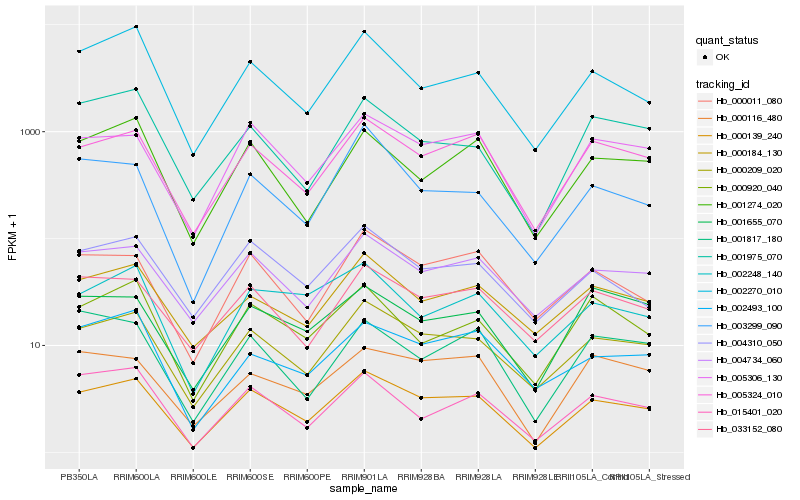
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_240 |
0.0 |
- |
- |
PREDICTED: putative F-box protein At3g16210 [Jatropha curcas] |
| 2 |
Hb_000209_020 |
0.0887592295 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 3 |
Hb_005324_010 |
0.0993141887 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 4 |
Hb_005306_130 |
0.1001695887 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_002270_010 |
0.1176883841 |
- |
- |
Chaperone protein dnaJ, putative [Ricinus communis] |
| 6 |
Hb_000011_080 |
0.1226377374 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 7 |
Hb_004734_060 |
0.1247519761 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 8 |
Hb_015401_020 |
0.1254841446 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_001274_020 |
0.1266959319 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter GCN1 [Hevea brasiliensis] |
| 10 |
Hb_000920_040 |
0.128713305 |
- |
- |
PREDICTED: F-box protein At1g30790-like [Jatropha curcas] |
| 11 |
Hb_000116_480 |
0.1292245899 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 12 |
Hb_003299_090 |
0.1309099765 |
- |
- |
hypothetical protein PRUPE_ppa011647mg [Prunus persica] |
| 13 |
Hb_004310_050 |
0.1341165591 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PK [Jatropha curcas] |
| 14 |
Hb_033152_080 |
0.1359454592 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 15 |
Hb_002248_140 |
0.1371517481 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |
| 16 |
Hb_002493_100 |
0.1384176584 |
- |
- |
PREDICTED: receptor-like protein kinase BRI1-like 3 [Jatropha curcas] |
| 17 |
Hb_000184_130 |
0.1392920222 |
- |
- |
PREDICTED: uncharacterized protein LOC105641544 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001817_180 |
0.139646794 |
- |
- |
PREDICTED: RING-H2 finger protein ATL47 [Jatropha curcas] |
| 19 |
Hb_001975_070 |
0.1402301735 |
- |
- |
eukaryotic translation elongation factor 1B alpha-subunit [Hevea brasiliensis] |
| 20 |
Hb_001655_070 |
0.1402886429 |
- |
- |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |