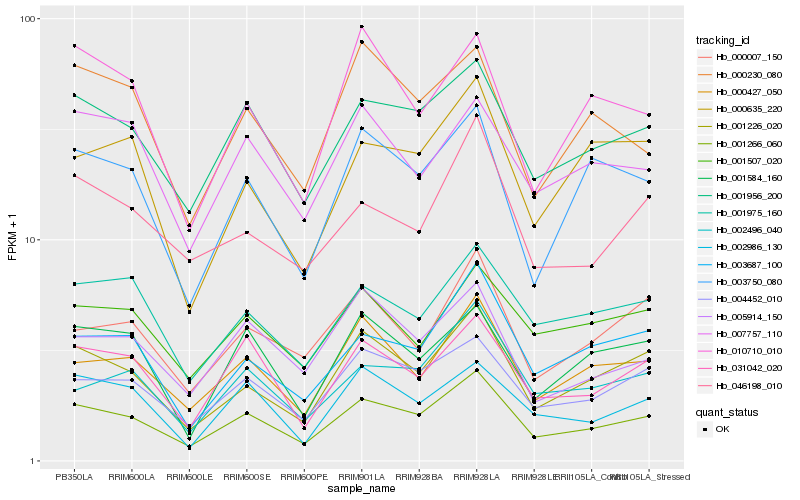
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001266_060 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g47840 [Jatropha curcas] |
| 2 |
Hb_001226_020 |
0.0723264464 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial-like [Jatropha curcas] |
| 3 |
Hb_001956_200 |
0.07391159 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_004452_010 |
0.0790898198 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 5 |
Hb_000427_050 |
0.0843238812 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_031042_020 |
0.0852983872 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g47460 [Jatropha curcas] |
| 7 |
Hb_003687_100 |
0.0932621731 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g23330 [Jatropha curcas] |
| 8 |
Hb_003750_080 |
0.0954429079 |
- |
- |
PREDICTED: DNA repair protein complementing XP-C cells homolog [Jatropha curcas] |
| 9 |
Hb_002496_040 |
0.0998938817 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g03380, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001975_160 |
0.1006935462 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002986_130 |
0.1038878181 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein DOT4, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_046198_010 |
0.1042172147 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 13 |
Hb_000007_150 |
0.1055596301 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g41080 [Jatropha curcas] |
| 14 |
Hb_005914_150 |
0.1064850956 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 15 |
Hb_001584_160 |
0.1082903102 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g56570 [Jatropha curcas] |
| 16 |
Hb_001507_020 |
0.1114654916 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14170 [Jatropha curcas] |
| 17 |
Hb_000635_220 |
0.1119918405 |
- |
- |
PREDICTED: F-box protein At1g67340 [Jatropha curcas] |
| 18 |
Hb_007757_110 |
0.1131475158 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5 [Jatropha curcas] |
| 19 |
Hb_000230_080 |
0.1132327724 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 20 |
Hb_010710_010 |
0.1133072817 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |