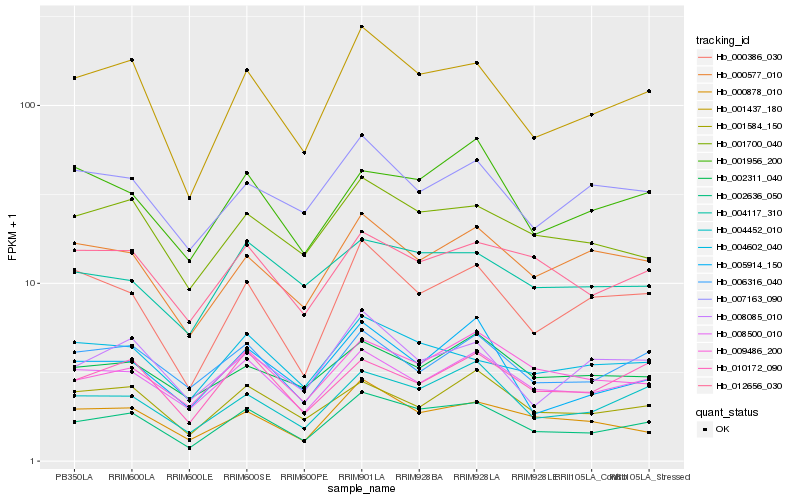
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002636_050 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g68930 [Jatropha curcas] |
| 2 |
Hb_001437_180 |
0.0562443386 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 3 |
Hb_004452_010 |
0.0850136179 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g58590 [Jatropha curcas] |
| 4 |
Hb_008500_010 |
0.0903223917 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17210 [Jatropha curcas] |
| 5 |
Hb_001700_040 |
0.0952093087 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 6 |
Hb_009486_200 |
0.095629671 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g09900-like [Jatropha curcas] |
| 7 |
Hb_001584_150 |
0.1001394603 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g40405 [Jatropha curcas] |
| 8 |
Hb_012656_030 |
0.1007626958 |
transcription factor |
TF Family: PHD |
PREDICTED: protein CHROMATIN REMODELING 4 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000386_030 |
0.1008620892 |
- |
- |
PREDICTED: uncharacterized protein LOC105629270 [Jatropha curcas] |
| 10 |
Hb_000878_010 |
0.1020647619 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g06400, mitochondrial [Jatropha curcas] |
| 11 |
Hb_004602_040 |
0.1022570141 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 12 |
Hb_000577_010 |
0.1035728997 |
- |
- |
hypothetical protein JCGZ_23887 [Jatropha curcas] |
| 13 |
Hb_010172_090 |
0.1042859651 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_005914_150 |
0.1052382505 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g19890 [Jatropha curcas] |
| 15 |
Hb_004117_310 |
0.106179414 |
- |
- |
PREDICTED: uncharacterized protein LOC105648043 [Jatropha curcas] |
| 16 |
Hb_006316_040 |
0.1064312629 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g16860 [Jatropha curcas] |
| 17 |
Hb_008085_010 |
0.1070864748 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g26680, mitochondrial [Jatropha curcas] |
| 18 |
Hb_001956_200 |
0.1110713694 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_002311_040 |
0.1116540621 |
- |
- |
hypothetical protein JCGZ_18966 [Jatropha curcas] |
| 20 |
Hb_007163_090 |
0.1128977944 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |