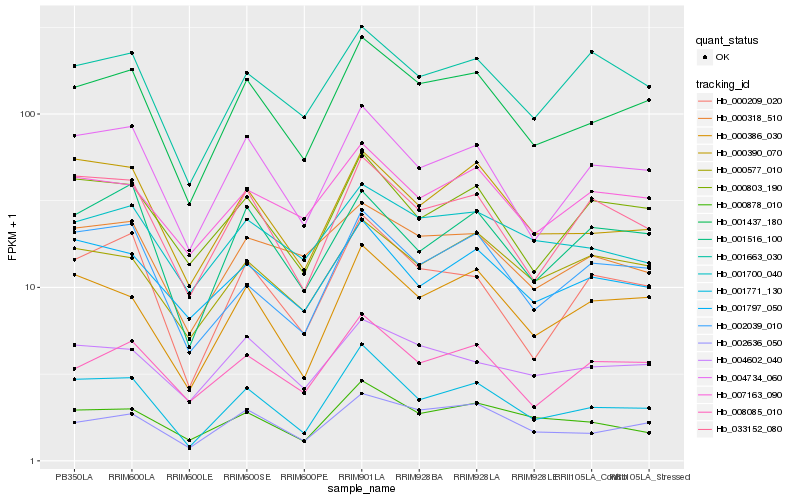
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001437_180 |
0.0 |
desease resistance |
Gene Name: CDC48_N |
PREDICTED: cell division cycle protein 48 homolog [Jatropha curcas] |
| 2 |
Hb_002636_050 |
0.0562443386 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g68930 [Jatropha curcas] |
| 3 |
Hb_004734_060 |
0.0749152606 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001771_130 |
0.0763977783 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g27610 [Jatropha curcas] |
| 5 |
Hb_008085_010 |
0.0816522817 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g26680, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000386_030 |
0.0823032781 |
- |
- |
PREDICTED: uncharacterized protein LOC105629270 [Jatropha curcas] |
| 7 |
Hb_004602_040 |
0.0932031285 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_000803_190 |
0.0938251557 |
- |
- |
PREDICTED: THUMP domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000318_510 |
0.0952745502 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15980 [Jatropha curcas] |
| 10 |
Hb_000390_070 |
0.0954734883 |
- |
- |
PREDICTED: uncharacterized protein LOC105632478 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001700_040 |
0.0955269132 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex non-core subunit NAF1 [Jatropha curcas] |
| 12 |
Hb_000209_020 |
0.095677048 |
- |
- |
PREDICTED: uncharacterized protein At4g26450 isoform X3 [Jatropha curcas] |
| 13 |
Hb_033152_080 |
0.0960041551 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 14 |
Hb_007163_090 |
0.0969190554 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 15 |
Hb_000577_010 |
0.0978024189 |
- |
- |
hypothetical protein JCGZ_23887 [Jatropha curcas] |
| 16 |
Hb_001663_030 |
0.1001604947 |
- |
- |
PREDICTED: peroxisomal membrane protein 13 [Jatropha curcas] |
| 17 |
Hb_001516_100 |
0.1004479225 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 18 |
Hb_002039_010 |
0.1006226328 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 19 |
Hb_000878_010 |
0.1016935337 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g06400, mitochondrial [Jatropha curcas] |
| 20 |
Hb_001797_050 |
0.1023290031 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 3-like isoform X1 [Jatropha curcas] |