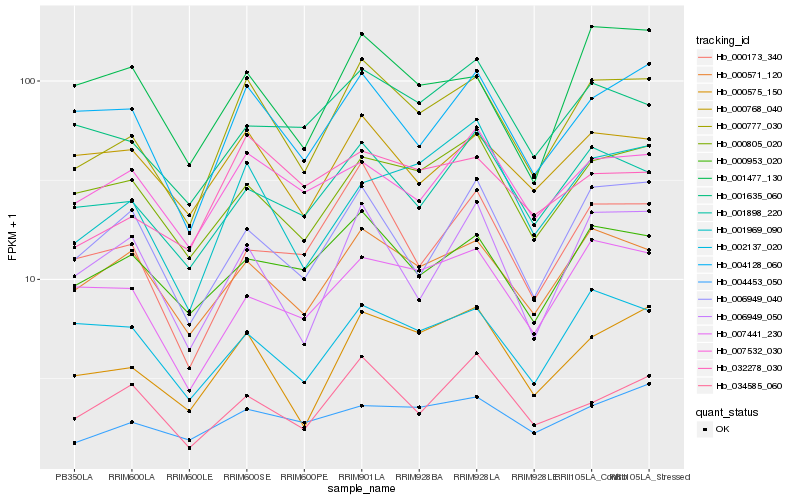
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000777_030 |
0.0 |
- |
- |
histone h2a, putative [Ricinus communis] |
| 2 |
Hb_000575_150 |
0.0922835155 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 3 |
Hb_000571_120 |
0.1000894548 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 4 |
Hb_004128_060 |
0.1075432118 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit A [Jatropha curcas] |
| 5 |
Hb_001898_220 |
0.1096992684 |
- |
- |
PREDICTED: regulator of nonsense transcripts UPF3-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000953_020 |
0.1124167233 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 7 |
Hb_000805_020 |
0.1131668559 |
- |
- |
pumilio/Puf RNA-binding domain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_032278_030 |
0.1186481106 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 9 |
Hb_006949_040 |
0.1191656028 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 10 |
Hb_000768_040 |
0.1195431648 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 11 |
Hb_007441_230 |
0.1219609032 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase [Jatropha curcas] |
| 12 |
Hb_001969_090 |
0.1223624911 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 13 |
Hb_000173_340 |
0.1224276312 |
- |
- |
hypothetical protein JCGZ_02075 [Jatropha curcas] |
| 14 |
Hb_006949_050 |
0.122824744 |
- |
- |
unknown [Lotus japonicus] |
| 15 |
Hb_004453_050 |
0.1229303395 |
- |
- |
PREDICTED: uncharacterized protein LOC105637675 [Jatropha curcas] |
| 16 |
Hb_007532_030 |
0.1246796685 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g30090 [Jatropha curcas] |
| 17 |
Hb_001477_130 |
0.1257152475 |
- |
- |
PREDICTED: uncharacterized protein LOC105646871 [Jatropha curcas] |
| 18 |
Hb_002137_020 |
0.1264792335 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 19 |
Hb_001635_060 |
0.1267438887 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 20 |
Hb_034585_060 |
0.1272949634 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g50420 [Jatropha curcas] |