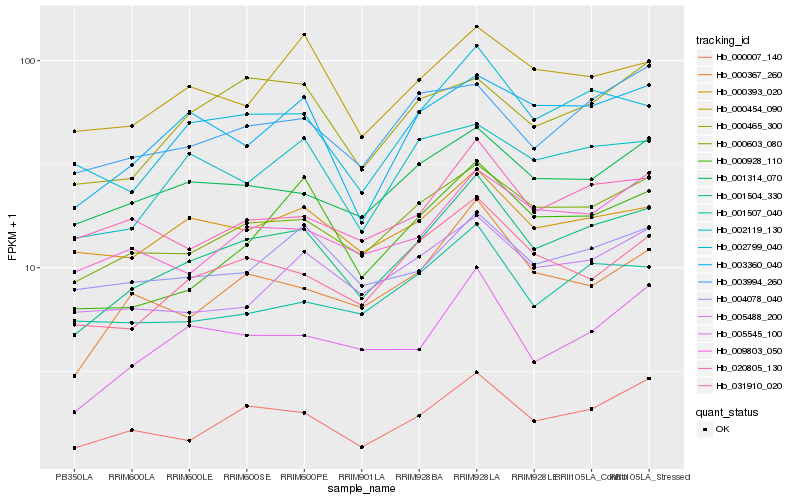
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_330 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 2 |
Hb_000603_080 |
0.0724417913 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170 [Vitis vinifera] |
| 3 |
Hb_002799_040 |
0.0796025087 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Vitis vinifera] |
| 4 |
Hb_000007_140 |
0.0820456436 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 5 |
Hb_031910_020 |
0.0880205581 |
- |
- |
PREDICTED: exocyst complex component SEC15A [Jatropha curcas] |
| 6 |
Hb_000367_260 |
0.0880626974 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 7 |
Hb_003360_040 |
0.0936626652 |
- |
- |
PREDICTED: uncharacterized protein LOC105634704 [Jatropha curcas] |
| 8 |
Hb_005545_100 |
0.0953188468 |
- |
- |
PREDICTED: uncharacterized protein LOC105645673 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001314_070 |
0.0977778031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002119_130 |
0.0981773205 |
- |
- |
PREDICTED: uncharacterized protein LOC105634169 [Jatropha curcas] |
| 11 |
Hb_009803_050 |
0.1007559467 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 12 |
Hb_000928_110 |
0.1015060438 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000393_020 |
0.1017389068 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 14 |
Hb_005488_200 |
0.1021902044 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000454_090 |
0.1024825056 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Tarenaya hassleriana] |
| 16 |
Hb_020805_130 |
0.1028434959 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_000465_300 |
0.1032235632 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 18 |
Hb_004078_040 |
0.1050486795 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 [Jatropha curcas] |
| 19 |
Hb_001507_040 |
0.1054249619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003994_260 |
0.1060201743 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |