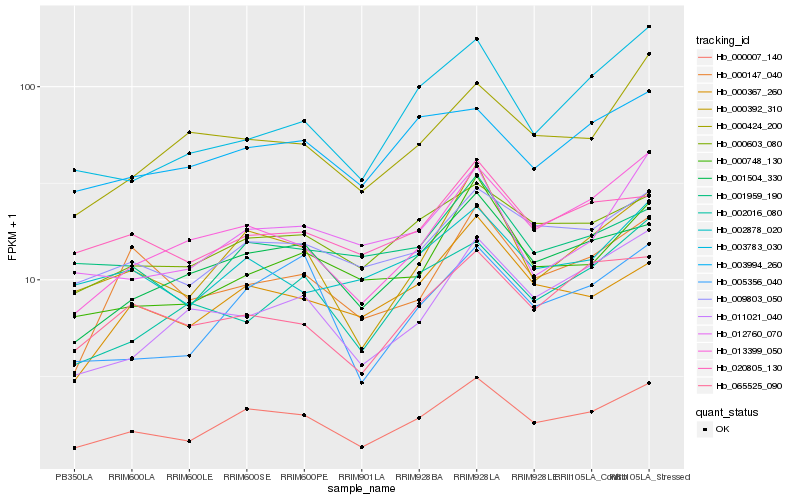
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_140 |
0.0 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 2 |
Hb_000392_310 |
0.0770999391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001504_330 |
0.0820456436 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 4 |
Hb_013399_050 |
0.0929609674 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_000603_080 |
0.0932107351 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170 [Vitis vinifera] |
| 6 |
Hb_009803_050 |
0.0932493704 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 7 |
Hb_005356_040 |
0.0970409052 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000748_130 |
0.1047008851 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000367_260 |
0.1047452057 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 10 |
Hb_065525_090 |
0.1101525929 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 11 |
Hb_003994_260 |
0.1129188338 |
- |
- |
PREDICTED: uncharacterized protein LOC105646151 [Jatropha curcas] |
| 12 |
Hb_000424_200 |
0.1135788567 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 13 |
Hb_020805_130 |
0.1152188818 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 14 |
Hb_011021_040 |
0.1154030872 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 15 |
Hb_002878_020 |
0.1155685733 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 16 |
Hb_001959_190 |
0.1156879925 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase family member SUVH9-like [Jatropha curcas] |
| 17 |
Hb_003783_030 |
0.1161117053 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 18 |
Hb_012760_070 |
0.1167165559 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_002016_080 |
0.1186067596 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000147_040 |
0.1186556502 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |