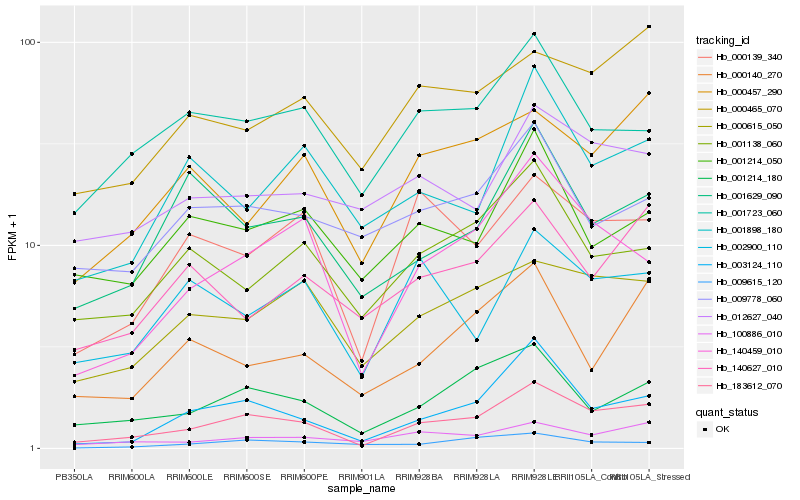
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183612_070 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_140459_010 |
0.133087717 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 3 |
Hb_003124_110 |
0.1399184283 |
- |
- |
hypothetical protein VITISV_026680 [Vitis vinifera] |
| 4 |
Hb_001214_180 |
0.145638983 |
- |
- |
- |
| 5 |
Hb_000615_050 |
0.1585497557 |
- |
- |
Potassium transporter 11 family protein [Populus trichocarpa] |
| 6 |
Hb_100886_010 |
0.1603040272 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_000139_340 |
0.1663062627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000465_070 |
0.1741584619 |
- |
- |
PREDICTED: 3-isopropylmalate dehydratase small subunit 3-like [Jatropha curcas] |
| 9 |
Hb_001138_060 |
0.1752636538 |
- |
- |
hypothetical protein CICLE_v10028086mg [Citrus clementina] |
| 10 |
Hb_001723_060 |
0.1765083827 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: LOW QUALITY PROTEIN: dihydrolipoyl dehydrogenase 2, mitochondrial [Populus euphratica] |
| 11 |
Hb_000457_290 |
0.1767027712 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 12 |
Hb_001629_090 |
0.1814408399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000140_270 |
0.1822522448 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 14 |
Hb_012627_040 |
0.1823102015 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 15 |
Hb_001898_180 |
0.1830693747 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_140627_010 |
0.1843441554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_009615_120 |
0.1855457953 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 18 |
Hb_001214_050 |
0.1855548007 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 19 |
Hb_009778_060 |
0.1866874919 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 20 |
Hb_002900_110 |
0.1871547348 |
- |
- |
amino acid transporter, putative [Ricinus communis] |