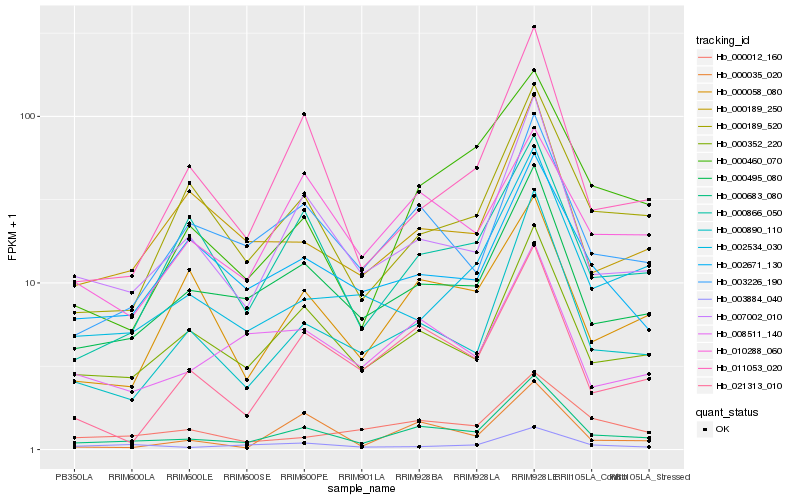
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000683_080 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 2 |
Hb_007002_010 |
0.1101430235 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 3 |
Hb_000495_080 |
0.1167970267 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 4 |
Hb_000352_220 |
0.1204471895 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000890_110 |
0.1209902012 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 6 |
Hb_003226_190 |
0.1445895054 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 7 |
Hb_000189_520 |
0.1528795089 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 8 |
Hb_021313_010 |
0.1536282467 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 9 |
Hb_011053_020 |
0.1549857388 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 10 |
Hb_000460_070 |
0.1614842303 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 11 |
Hb_000866_050 |
0.1623970317 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 12 |
Hb_000189_250 |
0.164285783 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002534_030 |
0.1645966641 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 14 |
Hb_008511_140 |
0.1646769235 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 15 |
Hb_000058_080 |
0.1672472586 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 16 |
Hb_000012_160 |
0.1716946296 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010288_060 |
0.1719749962 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 18 |
Hb_002671_130 |
0.173590746 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 19 |
Hb_000035_020 |
0.1749342912 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 20 |
Hb_003884_040 |
0.175891617 |
- |
- |
putative gag-pol polyprotein [Oryza sativa Japonica Group] |