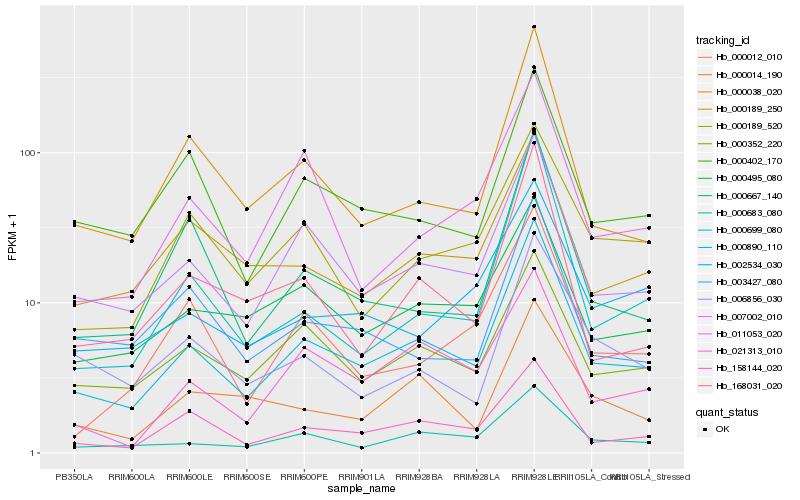
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_110 |
0.0 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 2 |
Hb_007002_010 |
0.0964018907 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 3 |
Hb_000683_080 |
0.1209902012 |
- |
- |
hypothetical protein EUGRSUZ_G013602, partial [Eucalyptus grandis] |
| 4 |
Hb_158144_020 |
0.124381527 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 5 |
Hb_000038_020 |
0.1280092793 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000352_220 |
0.1350592551 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 7 |
Hb_168031_020 |
0.1357520661 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000402_170 |
0.136224768 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 9 |
Hb_000495_080 |
0.1414552298 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 10 |
Hb_021313_010 |
0.1434168517 |
- |
- |
hypothetical protein POPTR_0010s24380g [Populus trichocarpa] |
| 11 |
Hb_000189_250 |
0.1459609346 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000667_140 |
0.1466937364 |
- |
- |
PREDICTED: uridine kinase-like protein 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000699_080 |
0.1486775777 |
- |
- |
PREDICTED: rubisco accumulation factor 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003427_080 |
0.1502878861 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000189_520 |
0.1504436397 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 16 |
Hb_011053_020 |
0.15185385 |
- |
- |
lipoic acid synthetase, putative [Ricinus communis] |
| 17 |
Hb_000012_010 |
0.1521731366 |
- |
- |
hypothetical protein JCGZ_07709 [Jatropha curcas] |
| 18 |
Hb_002534_030 |
0.1527844502 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 19 |
Hb_000014_190 |
0.1528316138 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_006856_030 |
0.155209399 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |