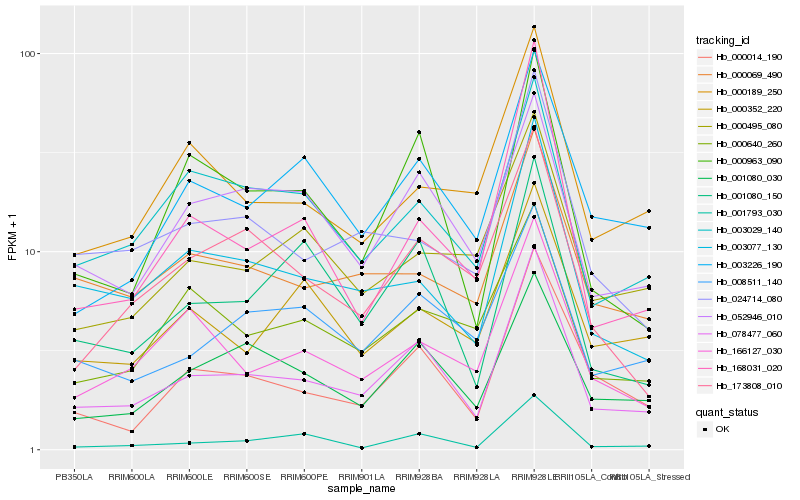
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_078477_060 |
0.0 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 2 |
Hb_000014_190 |
0.1087698977 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000963_090 |
0.119299223 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_003077_130 |
0.1235512614 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 5 |
Hb_001080_030 |
0.1317774222 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 6 |
Hb_001080_150 |
0.1329262913 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 7 |
Hb_001793_030 |
0.133020474 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 8 |
Hb_024714_080 |
0.135285071 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 9 |
Hb_008511_140 |
0.1366965174 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105632314 [Jatropha curcas] |
| 10 |
Hb_003226_190 |
0.1367194197 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 11 |
Hb_166127_030 |
0.141650379 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_168031_020 |
0.1428457984 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 13 |
Hb_052946_010 |
0.1430132031 |
- |
- |
PREDICTED: uncharacterized protein LOC105639103 [Jatropha curcas] |
| 14 |
Hb_000069_490 |
0.1461899901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003029_140 |
0.1464794562 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |
| 16 |
Hb_173808_010 |
0.1476302539 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000189_250 |
0.1505581011 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000495_080 |
0.1526049822 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |
| 19 |
Hb_000352_220 |
0.1541911519 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000640_260 |
0.1543839023 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g31400, chloroplastic [Jatropha curcas] |