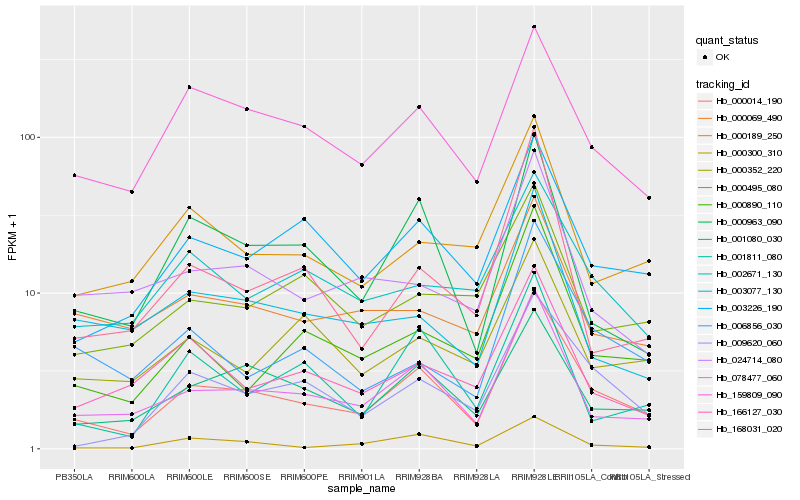
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000014_190 |
0.0 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_078477_060 |
0.1087698977 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 3 |
Hb_003226_190 |
0.1410355988 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 4 |
Hb_001080_030 |
0.1485777559 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |
| 5 |
Hb_009620_060 |
0.1488548608 |
- |
- |
PREDICTED: acetylornithine aminotransferase, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_000890_110 |
0.1528316138 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 7 |
Hb_000963_090 |
0.1578674741 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_159809_090 |
0.159040329 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-enyl diphosphate reductase [Hevea brasiliensis] |
| 9 |
Hb_000189_250 |
0.1635425006 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000300_310 |
0.1665796242 |
- |
- |
PREDICTED: uncharacterized protein LOC104896902 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_006856_030 |
0.1669259288 |
- |
- |
Uridine kinase-like protein 4 [Glycine soja] |
| 12 |
Hb_166127_030 |
0.1686294999 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_002671_130 |
0.1723004601 |
- |
- |
GTP-binding protein typa/bipa, putative [Ricinus communis] |
| 14 |
Hb_001811_080 |
0.1738784399 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 15 |
Hb_003077_130 |
0.174884861 |
- |
- |
hypothetical protein JCGZ_22476 [Jatropha curcas] |
| 16 |
Hb_000352_220 |
0.1759139546 |
- |
- |
PREDICTED: probable tyrosine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_024714_080 |
0.1763770864 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000069_490 |
0.1766027314 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_168031_020 |
0.1769876734 |
- |
- |
PREDICTED: uncharacterized protein LOC105646684 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000495_080 |
0.1774233287 |
- |
- |
Pentatricopeptide repeat (PPR-like) superfamily protein [Theobroma cacao] |