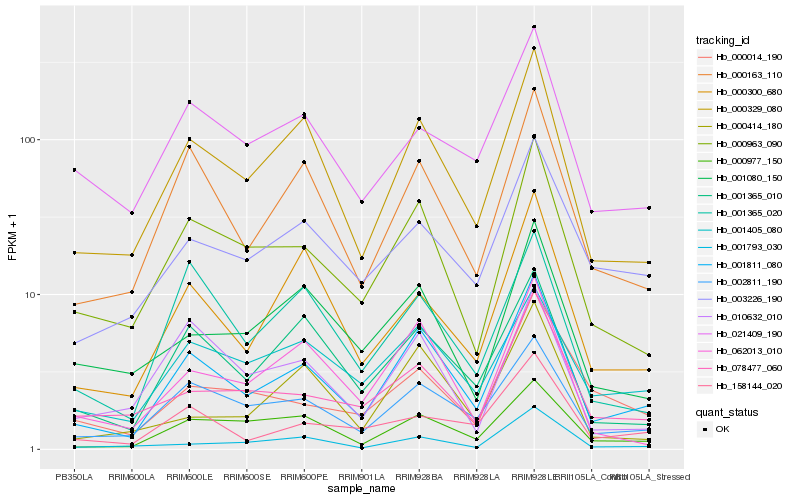
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001811_080 |
0.0 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 2 |
Hb_000329_080 |
0.1070360531 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002811_190 |
0.1153625067 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 4 |
Hb_000163_110 |
0.1180975194 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 5 |
Hb_000963_090 |
0.1229178795 |
rubber biosynthesis |
Gene Name: 4-hydroxy-3-methylbut-2-enyl diphosphate reductase |
4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase [Hevea brasiliensis] |
| 6 |
Hb_000977_150 |
0.1396015155 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 7 |
Hb_001365_010 |
0.1433769844 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 8 |
Hb_062013_010 |
0.1524612444 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_001405_080 |
0.1545493106 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 10 |
Hb_000300_680 |
0.1587833035 |
- |
- |
PREDICTED: uncharacterized protein LOC105632625 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003226_190 |
0.1594573159 |
- |
- |
PREDICTED: blue-light photoreceptor PHR2 [Jatropha curcas] |
| 12 |
Hb_078477_060 |
0.1595053636 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase ATX4 [Jatropha curcas] |
| 13 |
Hb_158144_020 |
0.1639458358 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Vitis vinifera] |
| 14 |
Hb_001793_030 |
0.1660664413 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 15 |
Hb_010632_010 |
0.1667472488 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_021409_190 |
0.1687212648 |
- |
- |
PREDICTED: luminal-binding protein 5-like [Jatropha curcas] |
| 17 |
Hb_001080_150 |
0.1698662581 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 18 |
Hb_001365_020 |
0.1699292914 |
- |
- |
hypothetical protein PRUPE_ppa000927mg [Prunus persica] |
| 19 |
Hb_000414_180 |
0.1732064831 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X2 [Vitis vinifera] |
| 20 |
Hb_000014_190 |
0.1738784399 |
- |
- |
Phytoene dehydrogenase, putative [Ricinus communis] |