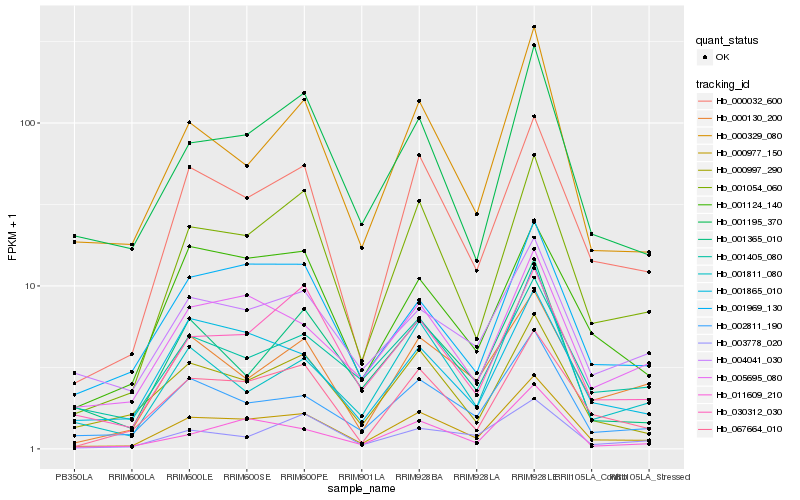
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000977_150 |
0.0 |
- |
- |
latex cyanogenic beta glucosidase [Hevea brasiliensis] |
| 2 |
Hb_000032_600 |
0.0919513586 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_001054_060 |
0.092583351 |
- |
- |
PREDICTED: dihydropyrimidine dehydrogenase [NADP(+)] [Jatropha curcas] |
| 4 |
Hb_002811_190 |
0.1025424513 |
- |
- |
tRNA pseudouridine synthase d, putative [Ricinus communis] |
| 5 |
Hb_067664_010 |
0.1036411341 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 6 |
Hb_000329_080 |
0.1102107175 |
- |
- |
PREDICTED: transketolase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001195_370 |
0.1161066092 |
- |
- |
- |
| 8 |
Hb_005695_080 |
0.1223364548 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 9 |
Hb_000997_290 |
0.1225590495 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 10 |
Hb_011609_210 |
0.1269765884 |
- |
- |
Uncharacterized protein TCM_011393 [Theobroma cacao] |
| 11 |
Hb_001969_130 |
0.1276188854 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |
| 12 |
Hb_030312_030 |
0.129686428 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000130_200 |
0.1298027553 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 14 |
Hb_001405_080 |
0.1323144212 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 15 |
Hb_001865_010 |
0.1330454968 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 16 |
Hb_004041_030 |
0.1383725932 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 17 |
Hb_001811_080 |
0.1396015155 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 18 |
Hb_003778_020 |
0.1432639503 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 19 |
Hb_001124_140 |
0.1448053898 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 20 |
Hb_001365_010 |
0.144833172 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |